HIV-1 Evolutionary Dynamics under Nonsuppressive Antiretroviral Therapy
- PMID: 35446121
- PMCID: PMC9239331
- DOI: 10.1128/mbio.00269-22
HIV-1 Evolutionary Dynamics under Nonsuppressive Antiretroviral Therapy
Abstract
Prolonged virologic failure on 2nd-line protease inhibitor (PI)-based antiretroviral therapy (ART) without emergence of major protease mutations is well recognized and provides an opportunity to study within-host evolution in long-term viremic individuals. Using next-generation sequencing and in silico haplotype reconstruction, we analyzed whole-genome sequences from longitudinal plasma samples of eight chronically infected HIV-1-positive individuals failing 2nd-line regimens from the French National Agency for AIDS and Viral Hepatitis Research (ANRS) 12249 Treatment as Prevention (TasP) trial. On nonsuppressive ART, there were large fluctuations in synonymous and nonsynonymous variant frequencies despite stable viremia. Reconstructed haplotypes provided evidence for selective sweeps during periods of partial adherence, and viral haplotype competition, during periods of low drug exposure. Drug resistance mutations in reverse transcriptase (RT) were used as markers of viral haplotypes in the reservoir, and their distribution over time indicated recombination. We independently observed linkage disequilibrium decay, indicative of recombination. These data highlight dramatic changes in virus population structure that occur during stable viremia under nonsuppressive ART. IMPORTANCE HIV-1 infections are most commonly initiated with a single founder virus and are characterized by extensive inter- and intraparticipant genetic diversity. However, existing literature on HIV-1 intrahost population dynamics is largely limited to untreated infections, predominantly in subtype B-infected individuals. The manuscript characterizes viral population dynamics in long-term viremic treatment-experienced individuals, which has not been previously characterized. These data are particularly relevant for understanding HIV dynamics but can also be applied to other RNA viruses. With this unique data set we propose that the virus is highly unstable, and we have found compelling evidence of HIV-1 within-host viral diversification, recombination, and haplotype competition during nonsuppressive ART.
Keywords: antiretroviral resistance; clinical failure; drug resistance evolution; human immunodeficiency virus.
Conflict of interest statement
The authors declare a conflict of interest. R.K.G. has received ad hoc consulting fees from Gilead, ViiV and UMOVIS Lab.
Figures


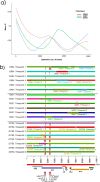



References
-
- Abrahams M-R, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping L-H, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, Center for HIV-AIDS Vaccine Immunology Consortium . 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. - DOI - PMC - PubMed
-
- Lemey P, Rambaut A, Pybus OG. 2006. HIV evolutionary dynamics within and among hosts. Aids Rev 8:125–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
