Integrated fecal microbiome-metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome
- PMID: 35446234
- PMCID: PMC9037519
- DOI: 10.1080/19490976.2022.2063016
Integrated fecal microbiome-metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome
Abstract
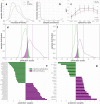
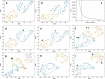
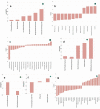
To gain insight into the complex microbiome-gut-brain axis in irritable bowel syndrome (IBS), several modalities of biological and clinical data must be combined. We aimed to identify profiles of fecal microbiota and metabolites associated with IBS and to delineate specific phenotypes of IBS that represent potential pathophysiological mechanisms. Fecal metabolites were measured using proton nuclear magnetic resonance (1H-NMR) spectroscopy and gut microbiome using shotgun metagenomic sequencing (MGS) in a combined dataset of 142 IBS patients and 120 healthy controls (HCs) with extensive clinical, biological and phenotype information. Data were analyzed using support vector classification and regression and kernel t-SNE. Microbiome and metabolome profiles could distinguish IBS and HC with an area-under-the-receiver-operator-curve of 77.3% and 79.5%, respectively, but this could be improved by combining microbiota and metabolites to 83.6%. No significant differences in predictive ability of the microbiome-metabolome data were observed between the three classical, stool pattern-based, IBS subtypes. However, unsupervised clustering showed distinct subsets of IBS patients based on fecal microbiome-metabolome data. These clusters could be related plasma levels of serotonin and its metabolite 5-hydroxyindoleacetate, effects of psychological stress on gastrointestinal (GI) symptoms, onset of IBS after stressful events, medical history of previous abdominal surgery, dietary caloric intake and IBS symptom duration. Furthermore, pathways in metabolic reaction networks were integrated with microbiota data, that reflect the host-microbiome interactions in IBS. The identified microbiome-metabolome signatures for IBS, associated with altered serotonin metabolism and unfavorable stress response related to GI symptoms, support the microbiota-gut-brain link in the pathogenesis of IBS.
Keywords: Irritable bowel syndrome; fecal metabolome; gut metabolome; gut microbiota; gut-brain; host-microbiome interaction; microbiome; serotonin; stress.
Conflict of interest statement
The authors have no conflicts of interest related to the content of this manuscript.With regard to any other disclosures: ZM holds the Niels Stensen Fellowship, Amsterdam. MK is supported by a Welcome Trust PhD Studentship in Basic Science (220119/Z/20/Z). DM partly financed by Grants of Top Knowledge Institute ‘Well on Wheat’ and ‘Well on Wheat 2.0’; the Carbokinetics program as part of the NOW CCC Partnership Program, H2020 nr 848228/DISCOvERIE and Organic A2BV/Mothersfinest BV IGP is supported by a National Institute for Health Research (NIHR) fellowship (NIHR-CDF-2017-10-032). The views expressed are those of the authors and not necessarily those of the UK National Health Service (NHS), the NIHR or the UK Department of Health. LV no conflicts of interest. ZW was supported by WillPharma to attend a scientific meeting. JISC is supported by the NIHR Imperial Biomedical Research Centre (BRC) in line with the Gut Health research theme. AZ is supported by the ERC Starting Grant 715772, Netherlands Organization for Scientific Research NWO-VIDI grant 016.178.056, and the Netherlands Heart Foundation CVON grant 2018-27. AZ and AK are supported by the NWO Gravitation grant ExposomeNL 024.004.017. JS no conflicts of interest. CW is supported by NWO Gravitation grant 024.003.001 and NWO Spinoza Prize SPI 92-266. EH is supported by a Premier’s Science Fellowship (Western Australia). DK has received research funding from Allergan, Will Pharma, Grunenthal, ZonMw, UEG, Rome Foundation, MLDS, outside of submitted work. JN acknowledges support from the Australian Federal Government. JP is supported by Health Data Research (HDR) UK and the Medical Research Council via a Rutherford Fund Fellowship (MR/S004033/1). AM has received a ZON MW, The Netherlands Organization for Health Research and Development, health care efficiency grant to evaluate efficacy of peppermint oil in IBS; has received an unrestricted research grant from Will Pharma S.A.; has received from the Dutch Cancer Society related to endoscopy and to colorectal polyps; received funding from Pentax Europe GmBH; received research funding from Allegan and Grünenthal on IBS topics; has given scientific advice to Bayer (topic: IBS) to Kyowa Kirin (topic: constipation) and to Takeda (topic: gastroparesis).
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
