Evolutionary changes between pre- and post-vaccine South African group A G2P[4] rotavirus strains, 2003-2017
- PMID: 35446251
- PMCID: PMC9453071
- DOI: 10.1099/mgen.0.000809
Evolutionary changes between pre- and post-vaccine South African group A G2P[4] rotavirus strains, 2003-2017
Abstract
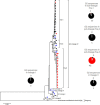
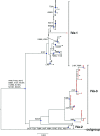
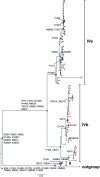
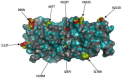
The transient upsurge of G2P[4] group A rotavirus (RVA) after Rotarix vaccine introduction in several countries has been a matter of concern. To gain insight into the diversity and evolution of G2P[4] strains in South Africa pre- and post-RVA vaccination introduction, whole-genome sequencing was performed for RVA positive faecal specimens collected between 2003 and 2017 and samples previously sequenced were obtained from GenBank (n=103; 56 pre- and 47 post-vaccine). Pre-vaccine G2 sequences predominantly clustered within sub-lineage IVa-1. In contrast, post-vaccine G2 sequences clustered mainly within sub-lineage IVa-3, whereby a radical amino acid (AA) substitution, S15F, was observed between the two sub-lineages. Pre-vaccine P[4] sequences predominantly segregated within sub-lineage IVa while post-vaccine sequences clustered mostly within sub-lineage IVb, with a radical AA substitution R162G. Both S15F and R162G occurred outside recognised antigenic sites. The AA residue at position 15 is found within the signal sequence domain of Viral Protein 7 (VP7) involved in translocation of VP7 into endoplasmic reticulum during infection process. The 162 AA residue lies within the hemagglutination domain of Viral Protein 4 (VP4) engaged in interaction with sialic acid-containing structure during attachment to the target cell. Free energy change analysis on VP7 indicated accumulation of stable point mutations in both antigenic and non-antigenic regions. The segregation of South African G2P[4] strains into pre- and post-vaccination sub-lineages is likely due to erstwhile hypothesized stepwise lineage/sub-lineage evolution of G2P[4] strains rather than RVA vaccine introduction. Our findings reinforce the need for continuous whole-genome RVA surveillance and investigation of contribution of AA substitutions in understanding the dynamic G2P[4] epidemiology.
Keywords: G2P[4] group A rotavirus strains; rotavirus; sub-lineages; whole-genome analysis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Estes MK, Greenberg HB. In: Fields Virology. 6th ed. Knipe DM, Howley PM, editors. Philadelphia (PA): Wolters Kluwer Heath/Lippincott Williams and Wilkins; 2013. Rotaviruses; pp. 1347–1401.
-
- Estes MK, Kapikian AZ, et al. In: Fields Virology. Knipe D, Griffin D, Lamb R, Martin M, Roizman B, editors. Philadelphia (PA): Wolters Kluwer Health/Lippincott Williams and Wilkins; 2007. Rotaviruses; pp. 1917–1975.
-
- Virus Classification Rega.kuleuven.be. 2021. [ December 13; 2021 ]. https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification accessed.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

