Riboswitch-inspired toehold riboregulators for gene regulation in Escherichia coli
- PMID: 35446427
- PMCID: PMC9071393
- DOI: 10.1093/nar/gkac275
Riboswitch-inspired toehold riboregulators for gene regulation in Escherichia coli
Abstract
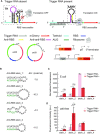
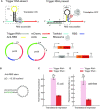
Regulatory RNA molecules have been widely investigated as components for synthetic gene circuits, complementing the use of protein-based transcription factors. Among the potential advantages of RNA-based gene regulators are their comparatively simple design, sequence-programmability, orthogonality, and their relatively low metabolic burden. In this work, we developed a set of riboswitch-inspired riboregulators in Escherichia coli that combine the concept of toehold-mediated strand displacement (TMSD) with the switching principles of naturally occurring transcriptional and translational riboswitches. Specifically, for translational activation and repression, we sequestered anti-anti-RBS or anti-RBS sequences, respectively, inside the loop of a stable hairpin domain, which is equipped with a single-stranded toehold region at its 5' end and is followed by regulated sequences on its 3' side. A trigger RNA binding to the toehold region can invade the hairpin, inducing a structural rearrangement that results in translational activation or deactivation. We also demonstrate that TMSD can be applied in the context of transcriptional regulation by switching RNA secondary structure involved in Rho-dependent termination. Our designs expand the repertoire of available synthetic riboregulators by a set of RNA switches with no sequence limitation, which should prove useful for the development of robust genetic sensors and circuits.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Nucleic acid strand displacement - from DNA nanotechnology to translational regulation.RNA Biol. 2023 Jan;20(1):154-163. doi: 10.1080/15476286.2023.2204565. RNA Biol. 2023. PMID: 37095744 Free PMC article.
-
Toehold switches: de-novo-designed regulators of gene expression.Cell. 2014 Nov 6;159(4):925-39. doi: 10.1016/j.cell.2014.10.002. Epub 2014 Oct 23. Cell. 2014. PMID: 25417166 Free PMC article.
-
Systematic Comparison and Rational Design of Theophylline Riboswitches for Effective Gene Repression.Microbiol Spectr. 2023 Feb 14;11(1):e0275222. doi: 10.1128/spectrum.02752-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688639 Free PMC article.
-
Developments of Riboswitches and Toehold Switches for Molecular Detection-Biosensing and Molecular Diagnostics.Int J Mol Sci. 2020 Apr 30;21(9):3192. doi: 10.3390/ijms21093192. Int J Mol Sci. 2020. PMID: 32366036 Free PMC article. Review.
-
Engineering Toehold-Mediated Switches for Native RNA Detection and Regulation in Bacteria.J Mol Biol. 2022 Sep 30;434(18):167689. doi: 10.1016/j.jmb.2022.167689. Epub 2022 Jun 16. J Mol Biol. 2022. PMID: 35717997 Review.
Cited by
-
Single-molecule force spectroscopy of toehold-mediated strand displacement.Nat Commun. 2024 Aug 31;15(1):7564. doi: 10.1038/s41467-024-51813-9. Nat Commun. 2024. PMID: 39217165 Free PMC article.
-
A CRISPR mis-insertion in the Zic3 5'UTR inhibits in vivo translation and is predicted to result in formation of an mRNA stem-loop hairpin.Biol Open. 2025 Mar 15;14(3):bio061677. doi: 10.1242/bio.061677. Epub 2025 Mar 17. Biol Open. 2025. PMID: 39912332 Free PMC article.
-
Design of a self-regulating mRNA gene circuit.Sci Rep. 2024 Aug 21;14(1):19421. doi: 10.1038/s41598-024-70363-0. Sci Rep. 2024. PMID: 39169208 Free PMC article.
-
Intelligent guide RNA: dual toehold switches for modulating luciferase in the presence of trigger RNA.Commun Biol. 2024 Oct 17;7(1):1344. doi: 10.1038/s42003-024-06988-8. Commun Biol. 2024. PMID: 39420075 Free PMC article.
-
Nucleic acid strand displacement - from DNA nanotechnology to translational regulation.RNA Biol. 2023 Jan;20(1):154-163. doi: 10.1080/15476286.2023.2204565. RNA Biol. 2023. PMID: 37095744 Free PMC article.
References
-
- Krützfeldt J., Stoffel M.. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006; 4:9–12. - PubMed
-
- Nudler E., Mironov A.S.. The riboswitch control of bacterial metabolism. Trends. Biochem. Sci. 2004; 29:11–17. - PubMed
-
- Amaral P.P., Mattick J.S.. Noncoding RNA in development. Mamm. Genome. 2008; 19:454–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

