The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update
- PMID: 35446428
- PMCID: PMC9252830
- DOI: 10.1093/nar/gkac247
The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update
Erratum in
-
Correction to 'The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update'.Nucleic Acids Res. 2022 Aug 26;50(15):8999. doi: 10.1093/nar/gkac610. Nucleic Acids Res. 2022. PMID: 35904799 Free PMC article. No abstract available.
Abstract
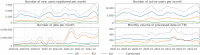
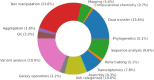
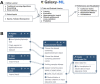
Galaxy is a mature, browser accessible workbench for scientific computing. It enables scientists to share, analyze and visualize their own data, with minimal technical impediments. A thriving global community continues to use, maintain and contribute to the project, with support from multiple national infrastructure providers that enable freely accessible analysis and training services. The Galaxy Training Network supports free, self-directed, virtual training with >230 integrated tutorials. Project engagement metrics have continued to grow over the last 2 years, including source code contributions, publications, software packages wrapped as tools, registered users and their daily analysis jobs, and new independent specialized servers. Key Galaxy technical developments include an improved user interface for launching large-scale analyses with many files, interactive tools for exploratory data analysis, and a complete suite of machine learning tools. Important scientific developments enabled by Galaxy include Vertebrate Genome Project (VGP) assembly workflows and global SARS-CoV-2 collaborations.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Schatz M.C., Philippakis A.A., Afgan E., Banks E., Carey V.J., Carroll R.J., Culotti A., Ellrott K., Goecks J., Grossman R.L.et al.. Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space. Cell Genomics. 2022; 2:100085. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

