Mitochondrial dynamics regulate genome stability via control of caspase-dependent DNA damage
- PMID: 35447090
- PMCID: PMC9616799
- DOI: 10.1016/j.devcel.2022.03.019
Mitochondrial dynamics regulate genome stability via control of caspase-dependent DNA damage
Abstract
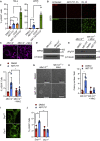
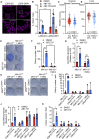
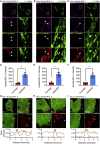
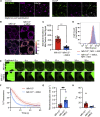
Mitochondrial dysfunction is interconnected with cancer. Nevertheless, how defective mitochondria promote cancer is poorly understood. We find that mitochondrial dysfunction promotes DNA damage under conditions of increased apoptotic priming. Underlying this process, we reveal a key role for mitochondrial dynamics in the regulation of DNA damage and genome instability. The ability of mitochondrial dynamics to regulate oncogenic DNA damage centers upon the control of minority mitochondrial outer membrane permeabilization (MOMP), a process that enables non-lethal caspase activation leading to DNA damage. Mitochondrial fusion suppresses minority MOMP and its associated DNA damage by enabling homogeneous mitochondrial expression of anti-apoptotic BCL-2 proteins. Finally, we find that mitochondrial dysfunction inhibits pro-apoptotic BAX retrotranslocation, causing BAX mitochondrial localization and thereby promoting minority MOMP. Unexpectedly, these data reveal oncogenic effects of mitochondrial dysfunction that are mediated via mitochondrial dynamics and caspase-dependent DNA damage.
Keywords: DNA damage; MOMP; apoptosis; cancer; caspase; cell death; fission; fusion; mitochondrial dynamics.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Berthenet K., Castillo Ferrer C., Fanfone D., Popgeorgiev N., Neves D., Bertolino P., Gibert B., Hernandez-Vargas H., Ichim G. Failed apoptosis enhances melanoma cancer cell aggressiveness. Cell Rep. 2020;31:107731. - PubMed
-
- Bock F.J., Tait S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020;21:85–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

