Biophysical Characterization of Membrane Proteins Embedded in Nanodiscs Using Fluorescence Correlation Spectroscopy
- PMID: 35448362
- PMCID: PMC9028781
- DOI: 10.3390/membranes12040392
Biophysical Characterization of Membrane Proteins Embedded in Nanodiscs Using Fluorescence Correlation Spectroscopy
Abstract
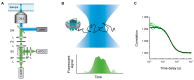
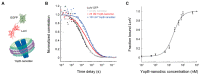
Proteins embedded in biological membranes perform essential functions in all organisms, serving as receptors, transporters, channels, cell adhesion molecules, and other supporting cellular roles. These membrane proteins comprise ~30% of all human proteins and are the targets of ~60% of FDA-approved drugs, yet their extensive characterization using established biochemical and biophysical methods has continued to be elusive due to challenges associated with the purification of these insoluble proteins. In response, the development of nanodisc techniques, such as nanolipoprotein particles (NLPs) and styrene maleic acid polymers (SMALPs), allowed membrane proteins to be expressed and isolated in solution as part of lipid bilayer rafts with defined, consistent nanometer sizes and compositions, thus enabling solution-based measurements. Fluorescence correlation spectroscopy (FCS) is a relatively simple yet powerful optical microscopy-based technique that yields quantitative biophysical information, such as diffusion kinetics and concentrations, about individual or interacting species in solution. Here, we first summarize current nanodisc techniques and FCS fundamentals. We then provide a focused review of studies that employed FCS in combination with nanodisc technology to investigate a handful of membrane proteins, including bacteriorhodopsin, bacterial division protein ZipA, bacterial membrane insertases SecYEG and YidC, Yersinia pestis type III secretion protein YopB, yeast cell wall stress sensor Wsc1, epidermal growth factor receptor (EGFR), ABC transporters, and several G protein-coupled receptors (GPCRs).
Keywords: cell-free expression; fluorescent correlation spectroscopy; membrane proteins; nanodiscs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- RCSB Protein Data Bank. [(accessed on 19 January 2022)]. Available online: https://www.rcsb.org/stats/growth/growth-protein.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

