Predictive Modeling of Alzheimer's and Parkinson's Disease Using Metabolomic and Lipidomic Profiles from Cerebrospinal Fluid
- PMID: 35448464
- PMCID: PMC9029812
- DOI: 10.3390/metabo12040277
Predictive Modeling of Alzheimer's and Parkinson's Disease Using Metabolomic and Lipidomic Profiles from Cerebrospinal Fluid
Abstract
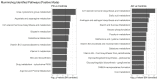
In recent years, metabolomics has been used as a powerful tool to better understand the physiology of neurodegenerative diseases and identify potential biomarkers for progression. We used targeted and untargeted aqueous, and lipidomic profiles of the metabolome from human cerebrospinal fluid to build multivariate predictive models distinguishing patients with Alzheimer's disease (AD), Parkinson's disease (PD), and healthy age-matched controls. We emphasize several statistical challenges associated with metabolomic studies where the number of measured metabolites far exceeds sample size. We found strong separation in the metabolome between PD and controls, as well as between PD and AD, with weaker separation between AD and controls. Consistent with existing literature, we found alanine, kynurenine, tryptophan, and serine to be associated with PD classification against controls, while alanine, creatine, and long chain ceramides were associated with AD classification against controls. We conducted a univariate pathway analysis of untargeted and targeted metabolite profiles and find that vitamin E and urea cycle metabolism pathways are associated with PD, while the aspartate/asparagine and c21-steroid hormone biosynthesis pathways are associated with AD. We also found that the amount of metabolite missingness varied by phenotype, highlighting the importance of examining missing data in future metabolomic studies.
Keywords: biomarker; cerebrospinal fluid; cross-sectional study; neurodegenerative disease; predictive modeling.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- Heron M. Deaths: Leading Causes for 2017. Natl. Vital. Stat. Rep. 2019;68:1–77. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

