NMR Metabolomics Reveal Urine Markers of Microbiome Diversity and Identify Benzoate Metabolism as a Mediator between High Microbial Alpha Diversity and Metabolic Health
- PMID: 35448495
- PMCID: PMC9025190
- DOI: 10.3390/metabo12040308
NMR Metabolomics Reveal Urine Markers of Microbiome Diversity and Identify Benzoate Metabolism as a Mediator between High Microbial Alpha Diversity and Metabolic Health
Abstract
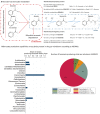
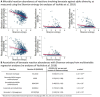
Microbial metabolites measured using NMR may serve as markers for physiological or pathological host-microbe interactions and possibly mediate the beneficial effects of microbiome diversity. Yet, comprehensive analyses of gut microbiome data and the urine NMR metabolome from large general population cohorts are missing. Here, we report the associations between gut microbiota abundances or metrics of alpha diversity, quantified from stool samples using 16S rRNA gene sequencing, with targeted urine NMR metabolites measures from 951 participants of the Study of Health in Pomerania (SHIP). We detected significant genus-metabolite associations for hippurate, succinate, indoxyl sulfate, and formate. Moreover, while replicating the previously reported association between hippurate and measures of alpha diversity, we identified formate and 4-hydroxyphenylacetate as novel markers of gut microbiome alpha diversity. Next, we predicted the urinary concentrations of each metabolite using genus abundances via an elastic net regression methodology. We found profound associations of the microbiome-based hippurate prediction score with markers of liver injury, inflammation, and metabolic health. Moreover, the microbiome-based prediction score for hippurate completely mediated the clinical association pattern of microbial diversity, hinting at a role of benzoate metabolism underlying the positive associations between high alpha diversity and healthy states. In conclusion, large-scale NMR urine metabolomics delivered novel insights into metabolic host-microbiome interactions, identifying pathways of benzoate metabolism as relevant candidates mediating the beneficial health effects of high microbial alpha diversity.
Keywords: NMR metabolomics; alpha diversity; benzoate metabolism; large cohort data; microbiome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. H.J.G. has received travel grants and speakers honoraria from Fresenius Medical Care, Neuraxpharm, Servier, and Janssen Cilag. A.P. has received lecturer honoraria from Technopath Clinical Diagnostics, Tosoh Bioscience, Roche Diagnostics, Beckmann Coulter GmbH, and Radiometer. M.N. has received travel grants by German Medical Association, German Centre for Cardiovascular Research, German Society for Clinical Chemistry and Laboratory Medicine, German National Cohort, German Research Foundation, Deutsche Akkreditierungsstelle, Sysmex, MDI Limbach, medpoint GmbH and Diasys; speaker’s honoraria from Novartis Pharma, Radiometer, AstraZeneca, Technopath Clinical Diagnostics, Sysmex, MDI Limbach, and medpoint Medizinkommunikations GmbH; and research funding from Aerocom GmbH and Profil Institut für Stoffwechselforschung GmbH.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

