The MicroRNA miR-277 Controls Physiology and Pathology of the Adult Drosophila Midgut by Regulating the Expression of Fatty Acid β-Oxidation-Related Genes in Intestinal Stem Cells
- PMID: 35448502
- PMCID: PMC9028014
- DOI: 10.3390/metabo12040315
The MicroRNA miR-277 Controls Physiology and Pathology of the Adult Drosophila Midgut by Regulating the Expression of Fatty Acid β-Oxidation-Related Genes in Intestinal Stem Cells
Abstract
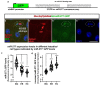
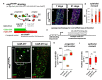
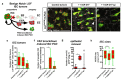
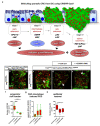
Cell division, growth, and differentiation are energetically costly and dependent processes. In adult stem cell-based epithelia, cellular identity seems to be coupled with a cell's metabolic profile and vice versa. It is thus tempting to speculate that resident stem cells have a distinct metabolism, different from more committed progenitors and differentiated cells. Although investigated for many stem cell types in vitro, in vivo data of niche-residing stem cell metabolism is scarce. In adult epithelial tissues, stem cells, progenitor cells, and their progeny have very distinct functions and characteristics. In our study, we hypothesized and tested whether stem and progenitor cell types might have a distinctive metabolic profile in the intestinal lineage. Here, taking advantage of the genetically accessible adult Drosophila melanogaster intestine and the availability of ex vivo single cell sequencing data, we tested that hypothesis and investigated the metabolism of the intestinal lineage from stem cell (ISC) to differentiated epithelial cell in their native context under homeostatic conditions. Our initial in silico analysis of single cell RNAseq data and functional experiments identify the microRNA miR-277 as a posttranscriptional regulator of fatty acid β-oxidation (FAO) in the intestinal lineage. Low levels of miR-277 are detected in ISC and progressively rising miR-277 levels are found in progenitors during their growth and differentiation. Supporting this, miR-277-regulated fatty acid β-oxidation enzymes progressively declined from ISC towards more differentiated cells in our pseudotime single-cell RNAseq analysis and in functional assays on RNA and protein level. In addition, in silico clustering of single-cell RNAseq data based on metabolic genes validates that stem cells and progenitors belong to two independent clusters with well-defined metabolic characteristics. Furthermore, studying FAO genes in silico indicates that two populations of ISC exist that can be categorized in mitotically active and quiescent ISC, of which the latter relies on FAO genes. In line with an FAO dependency of ISC, forced expression of miR-277 phenocopies RNAi knockdown of FAO genes by reducing ISC size and subsequently resulting in stem cell death. We also investigated miR-277 effects on ISC in a benign and our newly developed CRISPR-Cas9-based colorectal cancer model and found effects on ISC survival, which as a consequence affects tumor growth, further underlining the importance of FAO in a pathological context. Taken together, our study provides new insights into the basal metabolic requirements of intestinal stem cell on β-oxidation of fatty acids evolutionarily implemented by a sole microRNA. Gaining knowledge about the metabolic differences and dependencies affecting the survival of two central and cancer-relevant cell populations in the fly and human intestine might reveal starting points for targeted combinatorial therapy in the hope for better treatment of colorectal cancer in the future.
Keywords: Drosophila; fatty acid oxidation; intestinal stem cell; metabolism; midgut.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
HNF4 Regulates Fatty Acid Oxidation and Is Required for Renewal of Intestinal Stem Cells in Mice.Gastroenterology. 2020 Mar;158(4):985-999.e9. doi: 10.1053/j.gastro.2019.11.031. Epub 2019 Nov 22. Gastroenterology. 2020. PMID: 31759926 Free PMC article.
-
Fatty acid β-oxidation is required for the differentiation of larval hematopoietic progenitors in Drosophila.Elife. 2020 Jun 12;9:e53247. doi: 10.7554/eLife.53247. Elife. 2020. PMID: 32530419 Free PMC article.
-
Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut.Development. 2015 Feb 15;142(4):644-53. doi: 10.1242/dev.113357. Development. 2015. PMID: 25670791 Free PMC article.
-
Subversion of Niche-Signalling Pathways in Colorectal Cancer: What Makes and Breaks the Intestinal Stem Cell.Cancers (Basel). 2021 Feb 27;13(5):1000. doi: 10.3390/cancers13051000. Cancers (Basel). 2021. PMID: 33673710 Free PMC article. Review.
-
Intestinal stem cell response to injury: lessons from Drosophila.Cell Mol Life Sci. 2016 Sep;73(17):3337-49. doi: 10.1007/s00018-016-2235-9. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137186 Free PMC article. Review.
Cited by
-
Steroid hormone-induced wingless ligands tune female intestinal size in Drosophila.Nat Commun. 2025 Jan 6;16(1):436. doi: 10.1038/s41467-024-55664-2. Nat Commun. 2025. PMID: 39762218 Free PMC article.
-
miR-277 regulates the phase of circadian activity-rest rhythm in Drosophila melanogaster.Front Physiol. 2023 Nov 28;14:1082866. doi: 10.3389/fphys.2023.1082866. eCollection 2023. Front Physiol. 2023. PMID: 38089472 Free PMC article.
-
Identification of fatty acid oxidation-related subtypes by integrated analysis of bulk- and single-cell transcriptome profiling in colorectal cancer.J Gastrointest Oncol. 2024 Feb 29;15(1):147-163. doi: 10.21037/jgo-23-833. Epub 2024 Feb 23. J Gastrointest Oncol. 2024. PMID: 38482228 Free PMC article.
-
Noncoding RNA Regulation of Hormonal and Metabolic Systems in the Fruit Fly Drosophila.Metabolites. 2023 Jan 19;13(2):152. doi: 10.3390/metabo13020152. Metabolites. 2023. PMID: 36837772 Free PMC article. Review.
-
Inflammation- and Metastasis-Related Proteins Expression Changes in Early Stages in Tumor and Non-Tumor Adjacent Tissues of Colorectal Cancer Samples.Cancers (Basel). 2022 Sep 16;14(18):4487. doi: 10.3390/cancers14184487. Cancers (Basel). 2022. PMID: 36139645 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

