Adhesion as a Focus in Trichoderma-Root Interactions
- PMID: 35448603
- PMCID: PMC9026816
- DOI: 10.3390/jof8040372
Adhesion as a Focus in Trichoderma-Root Interactions
Abstract
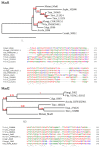
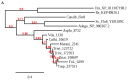
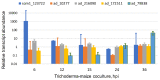
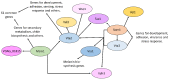
Fungal spores, germlings, and mycelia adhere to substrates, including host tissues. The adhesive forces depend on the substrate and on the adhesins, the fungal cell surface proteins. Attachment is often a prerequisite for the invasion of the host, hence its importance. Adhesion visibly precedes colonization of root surfaces and outer cortex layers, but little is known about the molecular details. We propose that by starting from what is already known from other fungi, including yeast and other filamentous pathogens and symbionts, the mechanism and function of Trichoderma adhesion will become accessible. There is a sequence, and perhaps functional, homology to other rhizosphere-competent Sordariomycetes. Specifically, Verticillium dahliae is a soil-borne pathogen that establishes itself in the xylem and causes destructive wilt disease. Metarhizium species are best-known as insect pathogens with biocontrol potential, but they also colonize roots. Verticillium orthologs of the yeast Flo8 transcription factor, Som1, and several other relevant genes are already under study for their roles in adhesion. Metarhizium encodes relevant adhesins. Trichoderma virens encodes homologs of Som1, as well as adhesin candidates. These genes should provide exciting leads toward the first step in the establishment of beneficial interactions with roots in the rhizosphere.
Keywords: Trichoderma; adhesin; adhesion; fungal; mutualism; rhizosphere; root; transcriptional.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mendoza-Mendoza A., Zaid R., Lawry R., Hermosa R., Monte E., Horwitz B.A., Mukherjee P.K. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Rev. 2018;32:62–85. doi: 10.1016/j.fbr.2017.12.001. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

