Benchmarking for biomedical natural language processing tasks with a domain specific ALBERT
- PMID: 35448946
- PMCID: PMC9022356
- DOI: 10.1186/s12859-022-04688-w
Benchmarking for biomedical natural language processing tasks with a domain specific ALBERT
Abstract
Background: The abundance of biomedical text data coupled with advances in natural language processing (NLP) is resulting in novel biomedical NLP (BioNLP) applications. These NLP applications, or tasks, are reliant on the availability of domain-specific language models (LMs) that are trained on a massive amount of data. Most of the existing domain-specific LMs adopted bidirectional encoder representations from transformers (BERT) architecture which has limitations, and their generalizability is unproven as there is an absence of baseline results among common BioNLP tasks.
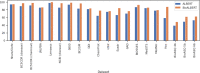
Results: We present 8 variants of BioALBERT, a domain-specific adaptation of a lite bidirectional encoder representations from transformers (ALBERT), trained on biomedical (PubMed and PubMed Central) and clinical (MIMIC-III) corpora and fine-tuned for 6 different tasks across 20 benchmark datasets. Experiments show that a large variant of BioALBERT trained on PubMed outperforms the state-of-the-art on named-entity recognition (+ 11.09% BLURB score improvement), relation extraction (+ 0.80% BLURB score), sentence similarity (+ 1.05% BLURB score), document classification (+ 0.62% F1-score), and question answering (+ 2.83% BLURB score). It represents a new state-of-the-art in 5 out of 6 benchmark BioNLP tasks.
Conclusions: The large variant of BioALBERT trained on PubMed achieved a higher BLURB score than previous state-of-the-art models on 5 of the 6 benchmark BioNLP tasks. Depending on the task, 5 different variants of BioALBERT outperformed previous state-of-the-art models on 17 of the 20 benchmark datasets, showing that our model is robust and generalizable in the common BioNLP tasks. We have made BioALBERT freely available which will help the BioNLP community avoid computational cost of training and establish a new set of baselines for future efforts across a broad range of BioNLP tasks.
Keywords: BioNLP; Bioinformatics; Biomedical text mining; Domain-specific language model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Storks S, Gao Q, Chai JY. Recent advances in natural language inference: a survey of benchmarks, resources, and approaches. 2019. arXiv:1904.01172.
-
- Peters M, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L. Deep contextualized word representations. In: Proceedings of the 2018 conference of the North American chapter of the association for computational linguistics: human language technologies, vol 1 (Long Papers). Association for Computational Linguistics; 2018, pp. 2227–2237. 10.18653/v1/N18-1202. http://aclweb.org/anthology/N18-1202.
-
- Devlin J, Chang M-W, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 conference of the North American chapter of the association for computational linguistics: human language technologies, vol 1 (long and short papers). 2019, pp. 4171–4186.
MeSH terms
LinkOut - more resources
Full Text Sources

