CMYA5 establishes cardiac dyad architecture and positioning
- PMID: 35449169
- PMCID: PMC9023524
- DOI: 10.1038/s41467-022-29902-4
CMYA5 establishes cardiac dyad architecture and positioning
Abstract
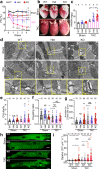
Cardiac excitation-contraction coupling requires dyads, the nanoscopic microdomains formed adjacent to Z-lines by apposition of transverse tubules and junctional sarcoplasmic reticulum. Disruption of dyad architecture and function are common features of diseased cardiomyocytes. However, little is known about the mechanisms that modulate dyad organization during cardiac development, homeostasis, and disease. Here, we use proximity proteomics in intact, living hearts to identify proteins enriched near dyads. Among these proteins is CMYA5, an under-studied striated muscle protein that co-localizes with Z-lines, junctional sarcoplasmic reticulum proteins, and transverse tubules in mature cardiomyocytes. During cardiac development, CMYA5 positioning adjacent to Z-lines precedes junctional sarcoplasmic reticulum positioning or transverse tubule formation. CMYA5 ablation disrupts dyad architecture, dyad positioning at Z-lines, and junctional sarcoplasmic reticulum Ca2+ release, leading to cardiac dysfunction and inability to tolerate pressure overload. These data provide mechanistic insights into cardiomyopathy pathogenesis by demonstrating that CMYA5 anchors junctional sarcoplasmic reticulum to Z-lines, establishes dyad architecture, and regulates dyad Ca2+ release.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

