Next generation automated traceless cell chromatography platform for GMP-compliant cell isolation and activation
- PMID: 35449227
- PMCID: PMC9023455
- DOI: 10.1038/s41598-022-10320-x
Next generation automated traceless cell chromatography platform for GMP-compliant cell isolation and activation
Abstract
Large-scale target cell isolation from patient blood preparations is one of the critical operations during drug product manufacturing for personalized cell therapy in immuno-oncology. Use of high-affinity murine antibody coated magnetic nanoparticles that remain on isolated cells is the current standard applied for this purpose. Here, we present the transformation of previously described technology - non-magnetic immunoaffinity column chromatography-based cell selection with reversible reagents into a new clinical-grade cell isolation platform called Automated Traceless Cell affinity chromatography (ATC). ATC is a fully closed and GMP-compliant cell selection and manufacturing system. Reversibility of reagents enables (sequential) positive cell selection, optionally in combination with depletion columns, enabling capture of highly specific cell subsets. Moreover, synergy with other Streptamer-based technologies allows novel uses beyond cell isolation including integrated and automated on-column target cell activation. In conclusion, ATC technology is an innovative as well as versatile platform to select, stimulate and modify cells for clinical manufacturing and downstream therapies.
© 2022. The Author(s).
Conflict of interest statement
S.R., M.P.P., M.W., V.C., C.R., I.T., S.P., A.W., S.A., B.B., E.B., S.D., F.F., G.N., N.H., T.S., D.H.B., M.E., S.P.F., C.S., and L.G. are currently employed by Juno Therapeutics GmbH, A Bristol-Myers Squibb Company and own stocks of Bristol-Myers Squibb. L.G., T.S., C.R., M.P.P., and C.S. are listed as inventors on previously filed related patent applications.
Figures
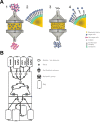
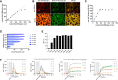



References
-
- Faraghat SA, Hoettges KF, Steinbach MK, van der Veen DR, Brackenbury WJ, Henslee EA, Labeed FH, Hughes MP. High-throughput, low-loss, low-cost, and label-free cell separation using electrophysiology-activated cell enrichment. Proc. Natl. Acad. Sci. USA. 2017;114(18):4591–4596. doi: 10.1073/pnas.1700773114. - DOI - PMC - PubMed
-
- Isozaki A, Mikami H, Tezuka H, Matsumura H, Huang K, Akamine M, Hiramatsu K, Iino T, Ito T, Karakawa H, Kasai Y, Li Y, Nakagawa Y, Ohnuki S, Ota T, Qian Y, Sakuma S, Sekiya T, Shirasaki Y, Suzuki N, Tayyabi E, Wakamiya T, Xu M, Yamagishi M, Yan H, Yu Q, Yan S, Yuan D, Zhang W, Zhao Y, Arai F, Campbell RE, Danelon C, Di Carlo D, Hiraki K, Hoshino Y, Hosokawa Y, Inaba M, Nakagawa A, Ohya Y, Oikawa M, Uemura S, Ozeki Y, Sugimura T, Nitta N, Goda K. Intelligent image-activated cell sorting 2.0. Lab Chip. 2020;20(13):2263–2273. doi: 10.1039/D0LC00080A. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

