Snapshots of actin and tubulin folding inside the TRiC chaperonin
- PMID: 35449234
- PMCID: PMC9113939
- DOI: 10.1038/s41594-022-00755-1
Snapshots of actin and tubulin folding inside the TRiC chaperonin
Abstract
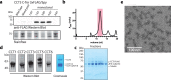
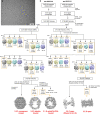
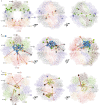
The integrity of a cell's proteome depends on correct folding of polypeptides by chaperonins. The chaperonin TCP-1 ring complex (TRiC) acts as obligate folder for >10% of cytosolic proteins, including he cytoskeletal proteins actin and tubulin. Although its architecture and how it recognizes folding substrates are emerging from structural studies, the subsequent fate of substrates inside the TRiC chamber is not defined. We trapped endogenous human TRiC with substrates (actin, tubulin) and cochaperone (PhLP2A) at different folding stages, for structure determination by cryo-EM. The already-folded regions of client proteins are anchored at the chamber wall, positioning unstructured regions toward the central space to achieve their native fold. Substrates engage with different sections of the chamber during the folding cycle, coupled to TRiC open-and-close transitions. Further, the cochaperone PhLP2A modulates folding, acting as a molecular strut between substrate and TRiC chamber. Our structural snapshots piece together an emerging model of client protein folding within TRiC.
© 2022. The Author(s).
Conflict of interest statement
K.M.K., J.M.J., and C.B. are employees of Pfizer Inc. J.J.K., D.T., E.P., G.C., H.K., L.H., J.S., V.O.P., J.T.H., and W.W.Y. declare no competing interests.
Figures













References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

