Genomes of leafy and leafless Platanthera orchids illuminate the evolution of mycoheterotrophy
- PMID: 35449401
- PMCID: PMC9023349
- DOI: 10.1038/s41477-022-01127-9
Genomes of leafy and leafless Platanthera orchids illuminate the evolution of mycoheterotrophy
Abstract
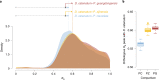
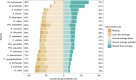
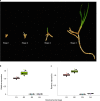
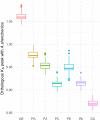
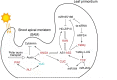
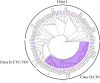
To improve our understanding of the origin and evolution of mycoheterotrophic plants, we here present the chromosome-scale genome assemblies of two sibling orchid species: partially mycoheterotrophic Platanthera zijinensis and holomycoheterotrophic Platanthera guangdongensis. Comparative analysis shows that mycoheterotrophy is associated with increased substitution rates and gene loss, and the deletion of most photoreceptor genes and auxin transporter genes might be linked to the unique phenotypes of fully mycoheterotrophic orchids. Conversely, trehalase genes that catalyse the conversion of trehalose into glucose have expanded in most sequenced orchids, in line with the fact that the germination of orchid non-endosperm seeds needs carbohydrates from fungi during the protocorm stage. We further show that the mature plant of P. guangdongensis, different from photosynthetic orchids, keeps expressing trehalase genes to hijack trehalose from fungi. Therefore, we propose that mycoheterotrophy in mature orchids is a continuation of the protocorm stage by sustaining the expression of trehalase genes. Our results shed light on the molecular mechanism underlying initial, partial and full mycoheterotrophy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




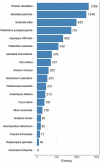
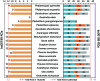
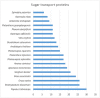
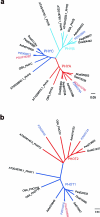



Comment in
-
Turning to the dark side.Nat Plants. 2022 Apr;8(4):324-325. doi: 10.1038/s41477-022-01132-y. Nat Plants. 2022. PMID: 35449402 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

