The Prognostic Signature of Head and Neck Squamous Cell Carcinoma Constructed by Immune-Related RNA-Binding Proteins
- PMID: 35449571
- PMCID: PMC9016149
- DOI: 10.3389/fonc.2022.795781
The Prognostic Signature of Head and Neck Squamous Cell Carcinoma Constructed by Immune-Related RNA-Binding Proteins
Abstract
Purpose: This study aimed to construct a prognostic signature consisting of immune-related RNA-binding proteins (RBPs) to predict the prognosis of patients with head and neck squamous cell carcinoma (HNSCC) effectively.
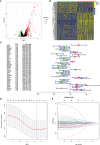
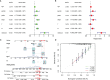
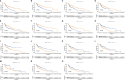
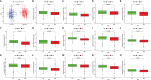
Methods: The transcriptome and clinical data of HNSCC were downloaded from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. First, we ascertained the immunological differences in HNSCC, through single-sample gene set enrichment analysis, stromal and immune cells in malignant tumor tissues using expression data (ESTIMATE), and cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT) deconvolution algorithm. Then we used univariate proportional hazards (Cox) regression analysis and least absolute shrinkage and selection operator (LASSO) Cox regression analysis to screen immune-related RBPs and acquire the risk score of each sample. Subsequently, we further investigated the difference in prognosis, immune status, and tumor mutation burden in high- and low-risk groups. Finally, the efficacy of immunotherapy was measured by the tumor immune dysfunction and exclusion (TIDE) score.
Results: We derived 15 immune-related RBPs, including FRMD4A, ASNS, RAB11FIP1, FAM120C, CFLAR, CTTN, PLEKHO1, SELENBP1, CHCHD2, NPM3, ATP2A3, CFDP1, IGF2BP2, NQO1, and DENND2D. There were significant differences in the prognoses of patients in the high- and low-risk groups in the training set (p < 0.001) and the validation set (p < 0.01). Furthermore, there were statistical differences between the high-risk group and low-risk group in immune cell infiltration and pathway and tumor mutation load (p < 0.001). In the end, we found that patients in the low-risk group were more sensitive to immunotherapy (p < 0.001), and then we screened 14 small-molecule chemotherapeutics with higher sensitivity to the high-risk group (p < 0.001).
Conclusion: The study constructed a prognostic signature of HNSCC, which might guide clinical immunotherapy in the future.
Keywords: RNA binding protein; chemotherapeutic; copy number variations; head and neck squamous cell carcinoma; immune microenvironment; immunotherapy; prognostic; tumor mutation burden.
Copyright © 2022 Ming, Li, Wang, Wei, Liu, Zhou, Yu, Zong and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
An angiogenesis-associated gene-based signature predicting prognosis and immunotherapy efficacy of head and neck squamous cell carcinoma patients.J Cancer Res Clin Oncol. 2024 Feb 12;150(2):91. doi: 10.1007/s00432-024-05606-8. J Cancer Res Clin Oncol. 2024. PMID: 38347320 Free PMC article.
-
The Prognostic Value of the DNA Repair Gene Signature in Head and Neck Squamous Cell Carcinoma.Front Oncol. 2021 Jul 30;11:710694. doi: 10.3389/fonc.2021.710694. eCollection 2021. Front Oncol. 2021. PMID: 34395285 Free PMC article.
-
A novel immunogenomic signature to predict prognosis and reveal immune infiltration characteristics in pancreatic ductal adenocarcinoma.Precis Clin Med. 2022 Apr 25;5(2):pbac010. doi: 10.1093/pcmedi/pbac010. eCollection 2022 Jun. Precis Clin Med. 2022. PMID: 35694712 Free PMC article.
-
Prognosis and Characterization of Immune Microenvironment in Head and Neck Squamous Cell Carcinoma through a Pyroptosis-Related Signature.J Oncol. 2022 Apr 6;2022:1539659. doi: 10.1155/2022/1539659. eCollection 2022. J Oncol. 2022. PMID: 35432539 Free PMC article.
-
A Narrative Review of Prognostic Gene Signatures in Oral Squamous Cell Carcinoma Using LASSO Cox Regression.Biomedicines. 2025 Jan 8;13(1):134. doi: 10.3390/biomedicines13010134. Biomedicines. 2025. PMID: 39857718 Free PMC article. Review.
Cited by
-
Transcriptomic signature associated with RNA-binding proteins for survival stratification of laryngeal cancer.Aging (Albany NY). 2022 Aug 18;14(16):6605-6625. doi: 10.18632/aging.204234. Epub 2022 Aug 18. Aging (Albany NY). 2022. PMID: 35985767 Free PMC article.
-
Identification of a new anoikis-related gene signature for prognostic significance in head and neck squamous carcinomas.Medicine (Baltimore). 2023 Sep 8;102(36):e34790. doi: 10.1097/MD.0000000000034790. Medicine (Baltimore). 2023. PMID: 37682196 Free PMC article.
-
TMCO1, as a potential biomarker of prognosis and immunotherapy response, regulates head and neck squamous cell carcinoma proliferation and migration.Discov Oncol. 2025 May 1;16(1):652. doi: 10.1007/s12672-025-02437-y. Discov Oncol. 2025. PMID: 40310585 Free PMC article.
-
T cell proliferation-related subtypes, prognosis model and characterization of tumor microenvironment in head and neck squamous cell carcinoma.Heliyon. 2024 Jul 10;10(14):e34221. doi: 10.1016/j.heliyon.2024.e34221. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39082023 Free PMC article.
-
Protein-RNA interactions: from mass spectrometry to drug discovery.Essays Biochem. 2023 Mar 29;67(2):175-186. doi: 10.1042/EBC20220177. Essays Biochem. 2023. PMID: 36866608 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

