In Silico Optimized Stapled Peptides Targeting WASF3 in Breast Cancer
- PMID: 35450347
- PMCID: PMC9014496
- DOI: 10.1021/acsmedchemlett.1c00627
In Silico Optimized Stapled Peptides Targeting WASF3 in Breast Cancer
Abstract
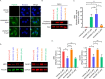
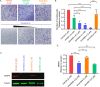
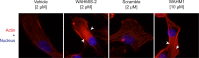
Wiskott-Aldrich Syndrome Protein Family (WASF) members regulate actin cytoskeletal dynamics, and WASF3 is directly associated with breast cancer metastasis and invasion. WASF3 forms a heteropentameric complex with CYFIP, NCKAP, ABI, and BRK1, called the WASF Regulatory Complex (WRC), which cooperatively regulates actin nucleation by WASF3. Since aberrant deployment of the WRC is observed in cancer metastasis and invasion, its disruption provides a novel avenue for targeting motility in breast cancer cells. Here, we report the development of a second generation WASF3 mimetic peptide, WAHMIS-2, which was designed using a combination of structure-guided design, homology modeling, and in silico optimization to disrupt binding of WASF3 to the WRC. WAHMIS-2 was found to permeate cells and inhibit cell motility, invasion, and MMP9 expression with greater potency than its predecessor, WAHM1. Targeted disruption of WASF3 from the WRC may serve as a useful strategy for suppression of breast cancer metastasis.
© 2022 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Targeting the WASF3 complex to suppress metastasis.Pharmacol Res. 2022 Aug;182:106302. doi: 10.1016/j.phrs.2022.106302. Epub 2022 Jun 9. Pharmacol Res. 2022. PMID: 35691539 Review.
-
Suppression of Breast Cancer Metastasis Using Stapled Peptides Targeting the WASF Regulatory Complex.Cancer Growth Metastasis. 2017 Jun 19;10:1179064417713197. doi: 10.1177/1179064417713197. eCollection 2017. Cancer Growth Metastasis. 2017. PMID: 28680267 Free PMC article.
-
Targeting the WASF3-CYFIP1 Complex Using Stapled Peptides Suppresses Cancer Cell Invasion.Cancer Res. 2016 Feb 15;76(4):965-73. doi: 10.1158/0008-5472.CAN-15-1680. Epub 2015 Dec 16. Cancer Res. 2016. PMID: 26676744 Free PMC article.
-
The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis.Cancer Res. 2016 Sep 1;76(17):5133-42. doi: 10.1158/0008-5472.CAN-16-0562. Epub 2016 Jul 18. Cancer Res. 2016. PMID: 27432794 Free PMC article.
-
Targeting WASF3 Signaling in Metastatic Cancer.Int J Mol Sci. 2021 Jan 15;22(2):836. doi: 10.3390/ijms22020836. Int J Mol Sci. 2021. PMID: 33467681 Free PMC article. Review.
Cited by
-
Discovering biomarkers associated and predicting cardiovascular disease with high accuracy using a novel nexus of machine learning techniques for precision medicine.Sci Rep. 2024 Jan 2;14(1):1. doi: 10.1038/s41598-023-50600-8. Sci Rep. 2024. PMID: 38167627 Free PMC article.
-
WASF3 overexpression affects the expression of circular RNA hsa-circ-0100153, which promotes breast cancer progression by sponging hsa-miR-31, hsa-miR-767-3p, and hsa-miR-935.Heliyon. 2023 Nov 29;9(12):e22874. doi: 10.1016/j.heliyon.2023.e22874. eCollection 2023 Dec. Heliyon. 2023. PMID: 38125536 Free PMC article.
-
WASP family proteins: Molecular mechanisms and implications in human disease.Eur J Cell Biol. 2022 Jun-Aug;101(3):151244. doi: 10.1016/j.ejcb.2022.151244. Epub 2022 Jun 1. Eur J Cell Biol. 2022. PMID: 35667337 Free PMC article.
-
The WAVE3/β-catenin oncogenic signaling regulates chemoresistance in triple negative breast cancer.Breast Cancer Res. 2023 Mar 22;25(1):31. doi: 10.1186/s13058-023-01634-3. Breast Cancer Res. 2023. PMID: 36949468 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

