Transcriptomic landscape of granulosa cells and peripheral blood mononuclear cells in women with PCOS compared to young poor responders and women with normal response
- PMID: 35451009
- PMCID: PMC9156844
- DOI: 10.1093/humrep/deac069
Transcriptomic landscape of granulosa cells and peripheral blood mononuclear cells in women with PCOS compared to young poor responders and women with normal response
Abstract
Study question: Are transcriptomic profiles altered in ovarian granulosa cells (GCs) and peripheral blood mononuclear cells (PBMNCs) of women with polycystic ovary syndrome (PCOS) compared to young poor responders (YPR) and women with normal response to ovarian stimulation?
Summary answer: RNA expression profiles in ovarian GCs and PBMNCs were significantly altered in patients with PCOS compared with normoresponder controls (CONT) and YPR.
What is known already: PCOS is characterised by a higher number of follicles at all developmental stages. During controlled ovarian hyperstimulation, PCOS women develop a larger number of follicles as a result of an exacerbated response, with an increased risk of ovarian hyperstimulation syndrome. Despite the number of developing follicles, they are often heterogeneous in both size and maturation stage, with compromised quality and retrieval of immature oocytes. Women with PCOS appear to have a longer reproductive lifespan, with a slightly higher menopausal age than the general population, in addition to having a higher antral follicular count. As a result, the ovarian follicular dynamics appear to differ significantly from those observed in women with poor ovarian response (POR) or diminished ovarian reserve.
Study design, size, duration: Transcriptomic profiling with RNA-sequencing and validation using quantitative reverse transcription PCR (qRT-PCR). Women with PCOS (N = 20), YPR (N = 20) and CONT (N = 20). Five patients for each group were used for sequencing and 15 samples per group were used for validation.
Participants/materials, setting, methods: PCOS was defined using the revised Rotterdam diagnostic criteria for PCOS. The YPR group included women <35 years old with <4 mature follicles (at least 15 mm) on the day of the trigger. According to internal data, this group represented the bottom 15th percentile of patients' responses in this age group. It was consistent with Patient-Oriented Strategies Encompassing Individualize D Oocyte Number (POSEIDON) criteria for POR (Group 3). The young CONT group included women <35 years without PCOS or anovulation, who developed >14 mature follicles (at least 15 mm on transvaginal ultrasound). According to internal data, a threshold of >14 mature follicles was established to represent the top 25% of patients in this age group in this clinic.Overall, n = 60 GCs and PBMNCs samples were collected and processed for total RNA extraction. To define the transcriptomic cargo of GCs and PBMNCs, RNA-seq libraries were successfully prepared from samples and analysed by RNA-seq analysis. Differential gene expression analysis was used to compare RNA-seq results between different groups of samples. Ingenuity pathway analysis was used to perform Gene Ontology and pathways analyses.
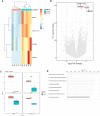
Main results and the role of chance: In PBMNCs of PCOS, there were 65 differentially expressed genes (DEGs) compared to CONT, and 16 compared to YPR. In GCs of PCOS, 4 genes showed decreased expression compared to CONT, while 58 genes were differentially expressed compared to YPR. qRT-PCR analysis confirmed the findings of the RNA-seq. The functional enrichment analysis performed revealed that DEGs in GCs of PCOS compared to CONT and YPR were prevalently involved in protein ubiquitination, oxidative phosphorylation, mitochondrial dysfunction and sirtuin signaling pathways.
Large scale data: The data used in this study is partially available at Gene Ontology database.
Limitations, reasons for caution: The analysis in PBMNCs could be uninformative due to inter-individual variability among patients in the same study groups. Despite the fact that we considered this was the best approach for our study's novel, exploratory nature.
Wider implications of the findings: RNA expression profiles in ovarian GCs and PBMNCs were altered in patients with PCOS compared with CONT and YPR. GCs of PCOS patients showed altered expression of several genes involved in oxidative phosphorylation, mitochondrial function and sirtuin signaling pathways. This is the first study to show that the transcriptomic landscape in GCs is altered in PCOS compared to CONT and YPR.
Study funding/competing interest(s): This study was partially supported by grant PI18/00322 from Instituto de Salud Carlos III, and European Regional Development Fund (FEDER), 'A way to make Europe' awarded to S.H. M.C., S.H., S.T., L.R., M.R., I.R., A.P. and R.C. declare no conflict of interests concerning this research. E.S. is a consultant for and receives research funding from the Foundation for Embryonic Competence.
Trial registration number: N/A.
Keywords: PCOS; mitochondrial dysfunction; oxidative phosphorylation; polycystic ovary syndrome; sirtuin pathway.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Figures



References
-
- Azziz R. Controversy in clinical endocrinology: diagnosis of polycystic ovarian syndrome: the Rotterdam criteria are premature. J Clin Endocrinol Metab 2006;91:781–785. - PubMed
-
- Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE et al. ; Task Force on the Phenotype of the Polycystic Ovary Syndrome of The Androgen Excess and PCOS Society. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril 2009;91:456–488. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300.
-
- Bhide P, Pundir J, Gudi A, Shah A, Homburg R, Acharya G. The effect of myo‐inositol/di‐chiro‐inositol on markers of ovarian reserve in women with PCOS undergoing IVF/ICSI: a systematic review and meta‐analysis. Acta Obstet Gynecol Scand 2019;98:1235–1244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

