Benchmarking statistical methods for analyzing parent-child dyads in genetic association studies
- PMID: 35451532
- PMCID: PMC9356976
- DOI: 10.1002/gepi.22453
Benchmarking statistical methods for analyzing parent-child dyads in genetic association studies
Abstract
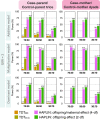
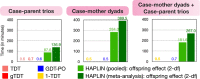
Genetic association studies of child health outcomes often employ family-based study designs. One of the most popular family-based designs is the case-parent trio design that considers the smallest possible nuclear family consisting of two parents and their affected child. This trio design is particularly advantageous for studying relatively rare disorders because it is less prone to type 1 error inflation due to population stratification compared to population-based study designs (e.g., case-control studies). However, obtaining genetic data from both parents is difficult, from a practical perspective, and many large studies predominantly measure genetic variants in mother-child dyads. While some statistical methods for analyzing parent-child dyad data (most commonly involving mother-child pairs) exist, it is not clear if they provide the same advantage as trio methods in protecting against population stratification, or if a specific dyad design (e.g., case-mother dyads vs. case-mother/control-mother dyads) is more advantageous. In this article, we review existing statistical methods for analyzing genome-wide marker data on dyads and perform extensive simulation experiments to benchmark their type I errors and statistical power under different scenarios. We extend our evaluation to existing methods for analyzing a combination of case-parent trios and dyads together. We apply these methods on genotyped and imputed data from multiethnic mother-child pairs only, case-parent trios only or combinations of both dyads and trios from the Gene, Environment Association Studies consortium (GENEVA), where each family was ascertained through a child affected by nonsyndromic cleft lip with or without cleft palate. Results from the GENEVA study corroborate the findings from our simulation experiments. Finally, we provide recommendations for using statistical genetic association methods for dyads.
Keywords: dyads; family-based GWAS; hybrid design; log-linear models; mother-child pairs; parent-offspring design; transmission disequilibrium; trios.
© 2022 The Authors. Genetic Epidemiology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Beaty, T. H. , Murray, J. C. , Marazita, M. L. , Munger, R. G. , Ruczinski, I. , Hetmanski, J. B. , Liang, K. Y. , Wu, T. , Murray, T. , Fallin, M. D. , Redett, R. A. , Raymond, G. , Schwender, H. , Jin, S. C. , Cooper, M. E. , Dunnwald, M. , Mansilla, M. A. , Leslie, E. , Bullard, S. , … Scott, A. F. (2010). A genome‐wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics, 42, 525–529. - PMC - PubMed
-
- Benyamin, B. , Visscher, P. M. , & McRae, A. F. (2009). Family‐based genome‐wide association studies. Pharmacogenomics, 10, 181–190. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

