Predictive functional assay-based classification of PMS2 variants in Lynch syndrome
- PMID: 35451539
- PMCID: PMC9545740
- DOI: 10.1002/humu.24387
Predictive functional assay-based classification of PMS2 variants in Lynch syndrome
Abstract
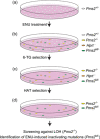
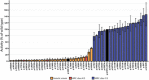
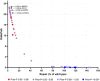
The large majority of germline alterations identified in the DNA mismatch repair (MMR) gene PMS2, a low-penetrance gene for the cancer predisposition Lynch syndrome, represent variants of uncertain significance (VUS). The inability to classify most VUS interferes with personalized healthcare. The complete in vitro MMR activity (CIMRA) assay, that only requires sequence information on the VUS, provides a functional analysis-based quantitative tool to improve the classification of VUS in MMR proteins. To derive a formula that translates CIMRA assay results into the odds of pathogenicity (OddsPath) for VUS in PMS2 we used a set of clinically classified PMS2 variants supplemented by inactivating variants that were generated by an in cellulo genetic screen, as proxies for cancer-predisposing variants. Validation of this OddsPath revealed high predictive values for benign and predisposing PMS2 VUS. We conclude that the OddsPath provides an integral metric that, following the other, higher penetrance, MMR proteins MSH2, MSH6 and MLH1 can be incorporated as strong evidence type into the upcoming criteria for MMR gene VUS classification of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP).
Keywords: DNA mismatch repair; Lynch syndrome; PMS2; diagnostic assessment; functional analysis-based classification; variants of uncertain significance.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Borràs, E. , Pineda, M. , Cadiñanos, J. , del Valle, J. , Brieger, A. , Hinrichsen, I. , & Capellá, G. (2013). Refining the role of PMS2 in Lynch syndrome: germline mutational analysis improved by comprehensive assessment of variants. J Journal of Medical Genetics, 50(8), 552–563. 10.1136/jmedgenet-2012-101511 - DOI - PubMed
-
- Brnich, S. E. , Abou Tayoun, A. N. , Couch, F. J. , Cutting, G. R. , Greenblatt, M. S. , Heinen, C. D. , Kanavy, D. M. , Luo, X. , McNulty, S. M. , Starita, L. M. , Tavtigian, S. V. , Wright, M. W. , Harrison, S. M. , Biesecker, L. G. , & Berg, J. S. , Clinical Genome Resource Sequence Variant Interpretation Working Group . (2019). Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Medicine, 12(1), 3. 10.1186/s13073-019-0690-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

