Assessment of T-cell Reactivity to the SARS-CoV-2 Omicron Variant by Immunized Individuals
- PMID: 35452102
- PMCID: PMC9034402
- DOI: 10.1001/jamanetworkopen.2022.10871
Assessment of T-cell Reactivity to the SARS-CoV-2 Omicron Variant by Immunized Individuals
Abstract
Importance: The emergence of the highly contagious Omicron variant of SARS-CoV-2 and the findings of a significantly reduced neutralizing potency of sera from individuals with previous SARS-CoV-2 infection or vaccination highlights the importance of studying cellular immunity to estimate the degree of immune protection to the new SARS-CoV-2 variant.
Objective: To determine T-cell reactivity to the Omicron variant in individuals with established (natural and/or vaccine-induced) immunity to SARS-CoV-2.
Design, setting, and participants: This was a cohort study conducted between December 20 and 21, 2021, at the Santa Lucia Foundation Istituto di Ricovero e Cura a Carattere Scientifico, Rome, Italy, among health care worker and scientist volunteers. Lymphocytes from freshly drawn blood samples were isolated and immediately tested for reactivity to the spike protein of SARS-CoV-2.
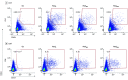
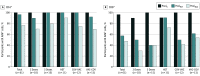
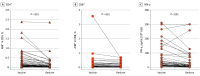
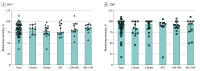
Main outcomes and measures: The main outcomes were the measurement of T-cell reactivity to the mutated regions of the spike protein of the Omicron BA.1 SARS-CoV-2 variant and the assessment of remaining T-cell immunity to the spike protein by stimulation with peptide libraries.
Results: A total of 61 volunteers (mean (range) age, 41.62 (21-62) years; 38 women [62%]) with different vaccination and SARS-CoV-2 infection backgrounds were enrolled. The median (range) frequency of CD4+ T cells reactive to peptides covering the mutated regions in the Omicron variant was 0.039% (0%-2.356%), a decrease of 64% compared with the frequency of CD4+ cells specific for the same regions of the ancestral strain (0.109% [0%-2.376%]). Within CD8+ T cells, a median (range) of 0.02% (0%-0.689%) of cells recognized the mutated spike regions, while 0.039% (0%-3.57%) of cells were reactive to the equivalent unmutated regions, a reduction of 49%. However, overall reactivity to the peptide library of the full-length protein was largely maintained (estimated 87%). No significant differences in loss of immune recognition were identified between groups of participants with different vaccination or infection histories.
Conclusions and relevance: This cohort study of immunized adults in Italy found that despite the mutations in the spike protein, the SARS-CoV-2 Omicron variant was recognized by the cellular component of the immune system. It is reasonable to assume that protection from hospitalization and severe disease will be maintained.
Conflict of interest statement
Figures
References
-
- World Health Organization . WHO coronavirus (COVID-19) dashboard. Accessed October 24, 2021. https://covid19.who.int/
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

