Structural and functional impact by SARS-CoV-2 Omicron spike mutations
- PMID: 35452593
- PMCID: PMC8995406
- DOI: 10.1016/j.celrep.2022.110729
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Abstract
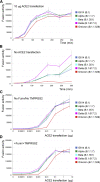
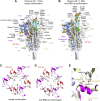
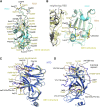
The Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), bearing an unusually high number of mutations, has become a dominant strain in many countries within several weeks. We report here structural, functional, and antigenic properties of its full-length spike (S) protein with a native sequence in comparison with those of previously prevalent variants. Omicron S requires a substantially higher level of host receptor ACE2 for efficient membrane fusion than other variants, possibly explaining its unexpected cellular tropism. Mutations not only remodel the antigenic structure of the N-terminal domain of the S protein but also alter the surface of the receptor-binding domain in a way not seen in other variants, consistent with its remarkable resistance to neutralizing antibodies. These results suggest that Omicron S has acquired an extraordinary ability to evade host immunity by excessive mutations, which also compromise its fusogenic capability.
Keywords: CP: Molecular biology; SARS-CoV-2; cryo-EM; spike protein; structure.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.Y. serves on the scientific advisory boards of Hummingbird Bioscience and GO Therapeutics and is currently an employee of GV20 Therapeutics LLC. All other authors declare no competing interests.
Figures


References
-
- Adamson B., Sikka R., Wyllie A.L., Premsrirut P. Discordant SARS-CoV-2 PCR and rapid antigen test results when infectious: a December 2021 occupational case series. medRxiv. 2022 doi: 10.1101/2022.01.04.22268770. Preprint at. - DOI
-
- Altarawneh H., Chemaitelly H., Tang P., Hasan M.R., Qassim S., Ayoub H.H., AlMukdad S., Yassine H.M., Benslimane F.M., Khatib H.A.A., et al. Protection afforded by prior infection against SARS-CoV-2 reinfection with the Omicron variant. medRxiv. 2022 doi: 10.1101/2022.01.05.22268782. Preprint at. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

