A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers
- PMID: 35452604
- PMCID: PMC9196205
- DOI: 10.1016/j.ccell.2022.03.012
A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers
Abstract
A common theme across multiple successful immunotherapies for cancer is the recognition of tumor-specific mutations (neoantigens) by T cells. The rapid discovery of such antigen responses could lead to improved therapies through the adoptive transfer of T cells engineered to express neoantigen-reactive T cell receptors (TCRs). Here, through CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) and TCR-seq of non-small cell lung cancer (NSCLC) tumor-infiltrating lymphocytes (TILs), we develop a neoantigen-reactive T cell signature based on clonotype frequency and CD39 protein and CXCL13 mRNA expression. Screening of TCRs selected by the signature allows us to identify neoantigen-reactive TCRs with a success rate of 45% for CD8+ and 66% for CD4+ T cells. Because of the small number of samples analyzed (4 patients), generalizability remains to be tested. However, this approach can enable the quick identification of neoantigen-reactive TCRs and expedite the engineering of personalized neoantigen-reactive T cells for therapy.
Keywords: CD39; CITE-seq; CXCL13; NSCLC; TILs; adoptive cell transfer therapy; neoantigens; single-cell analysis; tumor-infiltrating lymphocytes.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests. A non-provisional international patent has been filed based on the work described in this study.
Figures

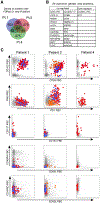



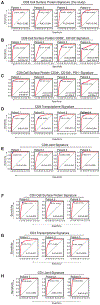


Comment in
-
Fast track to personalized TCR T cell therapies.Cancer Cell. 2022 May 9;40(5):447-449. doi: 10.1016/j.ccell.2022.04.013. Epub 2022 May 9. Cancer Cell. 2022. PMID: 35537408
References
-
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, Plaen ED, den Eynde BV, Knuth A, and Boon T (1991). A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

