Simultaneous brain cell type and lineage determined by scRNA-seq reveals stereotyped cortical development
- PMID: 35452605
- PMCID: PMC9233029
- DOI: 10.1016/j.cels.2022.03.006
Simultaneous brain cell type and lineage determined by scRNA-seq reveals stereotyped cortical development
Abstract
Mutations are acquired frequently, such that each cell's genome inscribes its history of cell divisions. Common genomic alterations involve loss of heterozygosity (LOH). LOH accumulates throughout the genome, offering large encoding capacity for inferring cell lineage. Using only single-cell RNA sequencing (scRNA-seq) of mouse brain cells, we found that LOH events spanning multiple genes are revealed as tracts of monoallelically expressed, constitutionally heterozygous single-nucleotide variants (SNVs). We simultaneously inferred cell lineage and marked developmental time points based on X chromosome inactivation and the total number of LOH events while identifying cell types from gene expression patterns. Our results are consistent with progenitor cells giving rise to multiple cortical cell types through stereotyped expansion and distinct waves of neurogenesis. This type of retrospective analysis could be incorporated into scRNA-seq pipelines and, compared with experimental approaches for determining lineage in model organisms, is applicable where genetic engineering is prohibited, such as humans.
Keywords: LOH; X-inactivation; brain development; cell fate; lineage; loss of heterozygosity.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


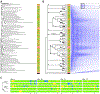
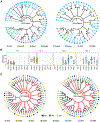

References
-
- Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics 43, 11.10.1–11.10.33. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

