Ribosome-associated quality-control mechanisms from bacteria to humans
- PMID: 35452614
- PMCID: PMC9034055
- DOI: 10.1016/j.molcel.2022.03.038
Ribosome-associated quality-control mechanisms from bacteria to humans
Abstract
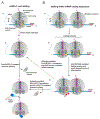
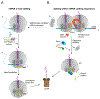
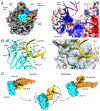
Ribosome-associated quality-control (RQC) surveys incomplete nascent polypeptides produced by interrupted translation. Central players in RQC are the human ribosome- and tRNA-binding protein, NEMF, and its orthologs, yeast Rqc2 and bacterial RqcH, which sense large ribosomal subunits obstructed with nascent chains and then promote nascent-chain proteolysis. In canonical eukaryotic RQC, NEMF stabilizes the LTN1/Listerin E3 ligase binding to obstructed ribosomal subunits for nascent-chain ubiquitylation. Furthermore, NEMF orthologs across evolution modify nascent chains by mediating C-terminal, untemplated polypeptide elongation. In eukaryotes, this process exposes ribosome-buried nascent-chain lysines, the ubiquitin acceptor sites, to LTN1. Remarkably, in both bacteria and eukaryotes, C-terminal tails also have an extra-ribosomal function as degrons. Here, we discuss recent findings on RQC mechanisms and briefly review how ribosomal stalling is sensed upstream of RQC, including via ribosome collisions, from an evolutionary perspective. Because RQC defects impair cellular fitness and cause neurodegeneration, this knowledge provides a framework for pathway-related biology and disease studies.
Keywords: C-terminal tail; CAT tail; KLHDC10; LTN1; Listerin; NEMF; Pirh2; RQC; Rqc2; RqcH; alanine tail; quality control; ribosome; ribosome collision; ribosome stalling; translation.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
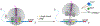

References
-
- Ahmed A, Wang M, Bergant G, Maroofian R, Zhao R, Alfadhel M, Nashabat M, AlRifai MT, Eyaid W, Alswaid A, et al. (2021). Biallelic loss-of-function variants in NEMF cause central nervous system impairment and axonal polyneuropathy. Hum. Genet 140, 579–592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

