Transcriptomic Analysis of E. coli after Exposure to a Sublethal Concentration of Hydrogen Peroxide Revealed a Coordinated Up-Regulation of the Cysteine Biosynthesis Pathway
- PMID: 35453340
- PMCID: PMC9026346
- DOI: 10.3390/antiox11040655
Transcriptomic Analysis of E. coli after Exposure to a Sublethal Concentration of Hydrogen Peroxide Revealed a Coordinated Up-Regulation of the Cysteine Biosynthesis Pathway
Abstract
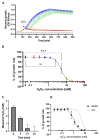
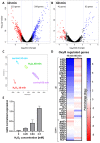
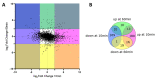
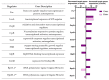
Hydrogen peroxide (H2O2) is a key defense component of host-microbe interaction. However, H2O2 concentrations generated by immune cells or epithelia are usually insufficient for bacterial killing and rather modulate bacterial responses. Here, we investigated the impact of sublethal H2O2 concentration on gene expression of E. coli BW25113 after 10 and 60 min of exposure. RNA-seq analysis revealed that approximately 12% of bacterial genes were strongly dysregulated 10 min following exposure to 2.5 mM H2O2. H2O2 exposure led to the activation of a specific antioxidant response and a general stress response. The latter was characterized by a transient down-regulation of genes involved in general metabolism, such as nucleic acid biosynthesis and translation, with a striking and coordinated down-regulation of genes involved in ribosome formation, and a sustained up-regulation of the SOS response. We confirmed the rapid transient and specific response mediated by the transcription factor OxyR leading to up-regulation of antioxidant systems, including the catalase-encoding gene (katG), that rapidly degrade extracellular H2O2 and promote bacterial survival. We documented a strong and transient up-regulation of genes involved in sulfur metabolism and cysteine biosynthesis, which are under the control of the transcription factor CysB. This strong specific transcriptional response to H2O2 exposure had no apparent impact on bacterial survival, but possibly replenishes the stores of oxidized cysteine and glutathione. In summary, our results demonstrate that different stress response mechanisms are activated by H2O2 exposure and highlight the cysteine synthesis as an antioxidant response in E. coli.
Keywords: E. coli; RNA-seq; cysB; cysteine biosynthesis; hydrogen peroxide; oxidative stress; sulfur assimilation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

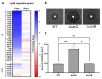
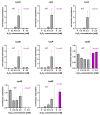
References
-
- Federal office of public health FOPH, Communicable Disease Division, Bern Switzerland: How Is Antibiotic Resistance Evolving? [(accessed on 5 November 2021)]. Available online: https://www.bag.admin.ch/bag/en/home/krankheiten/infektionskrankheiten-b....
-
- Tacconelli E. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organisation; Geneva, Switzerland: 2017. p. 7.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

