Novel User-Friendly Application for MRI Segmentation of Brain Resection following Epilepsy Surgery
- PMID: 35454065
- PMCID: PMC9032020
- DOI: 10.3390/diagnostics12041017
Novel User-Friendly Application for MRI Segmentation of Brain Resection following Epilepsy Surgery
Abstract
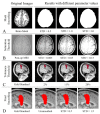
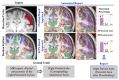
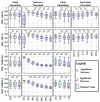
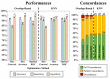
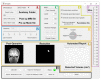
Delineation of resected brain cavities on magnetic resonance images (MRIs) of epilepsy surgery patients is essential for neuroimaging/neurophysiology studies investigating biomarkers of the epileptogenic zone. The gold standard to delineate the resection on MRI remains manual slice-by-slice tracing by experts. Here, we proposed and validated a semiautomated MRI segmentation pipeline, generating an accurate model of the resection and its anatomical labeling, and developed a graphical user interface (GUI) for user-friendly usage. We retrieved pre- and postoperative MRIs from 35 patients who had focal epilepsy surgery, implemented a region-growing algorithm to delineate the resection on postoperative MRIs and tested its performance while varying different tuning parameters. Similarity between our output and hand-drawn gold standards was evaluated via dice similarity coefficient (DSC; range: 0-1). Additionally, the best segmentation pipeline was trained to provide an automated anatomical report of the resection (based on presurgical brain atlas). We found that the best-performing set of parameters presented DSC of 0.83 (0.72-0.85), high robustness to seed-selection variability and anatomical accuracy of 90% to the clinical postoperative MRI report. We presented a novel user-friendly open-source GUI that implements a semiautomated segmentation pipeline specifically optimized to generate resection models and their anatomical reports from epilepsy surgery patients, while minimizing user interaction.
Keywords: MRI; brain resection; epilepsy surgery; image segmentation; region growing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Abdallah C., Maillard L.G., Rikir E., Jonas J., Thiriaux A., Gavaret M., Bartolomei F., Colnat-Coulbois S., Vignal J.P., Koessler L. Localizing Value of Electrical Source Imaging: Frontal Lobe, Malformations of Cortical Development and Negative MRI Related Epilepsies Are the Best Candidates. NeuroImage Clin. 2017;16:319–329. doi: 10.1016/j.nicl.2017.08.009. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

