In Silico Analysis of the Antagonist Effect of Enoxaparin on the ApoE4-Amyloid-Beta (A β) Complex at Different pH Conditions
- PMID: 35454088
- PMCID: PMC9027285
- DOI: 10.3390/biom12040499
In Silico Analysis of the Antagonist Effect of Enoxaparin on the ApoE4-Amyloid-Beta (A β) Complex at Different pH Conditions
Abstract
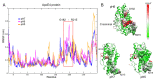
Apolipoprotein E4 (ApoE4) is thought to increase the risk of developing Alzheimer's disease. Several studies have shown that ApoE4-Amyloid β (Aβ) interactions can increment amyloid depositions in the brain and that this can be augmented at low pH values. On the other hand, experimental studies in transgenic mouse models have shown that treatment with enoxaparin significantly reduces cortical Aβ levels, as well as decreases the number of activated astrocytes around Aβ plaques. However, the interactions between enoxaparin and the ApoE4-Aβ proteins have been poorly explored. In this work, we combine molecular dynamics simulations, molecular docking, and binding free energy calculations to elucidate the molecular properties of the ApoE4-Aβ interactions and the competitive binding affinity of the enoxaparin on the ApoE4 binding sites. In addition, we investigated the effect of the environmental pH levels on those interactions. Our results showed that under different pH conditions, the closed form of the ApoE4 protein, in which the C-terminal domain folds into the protein, remains stabilized by a network of hydrogen bonds. This closed conformation allowed the generation of six different ApoE4-Aβ interaction sites, which were energetically favorable. Systems at pH5 and 6 showed the highest energetic affinity. The enoxaparin molecule was found to have a strong energetic affinity for ApoE4-interacting sites and thus can neutralize or disrupt ApoE4-Aβ complex formation.
Keywords: alzheimer disease; amyloid-β; apolipoprotein E; enoxaparin; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

