Genetic Diversity of Porcine Circovirus Types 2 and 3 in Wild Boar in Italy
- PMID: 35454199
- PMCID: PMC9031215
- DOI: 10.3390/ani12080953
Genetic Diversity of Porcine Circovirus Types 2 and 3 in Wild Boar in Italy
Abstract
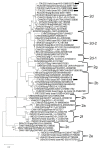
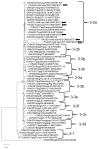
Porcine circovirus (PCV) infection is associated with relevant economic impact to the pig industry. To date, four species of PCV (PCV1 to 4) have been identified but only PCV2 has been associated firmly with disease in pigs. The objective of this study was to assess the prevalence of PCV2 and PCV3 in the wild boar population in Basilicata region, Southern Italy, since this region is characterized by large forested and rural areas and the anthropic pressure is lower than in other Italian regions. Liver samples from 82 hunted wild boar were collected in 2021 from 3 different hunting districts. Sixty (73%, 95%CI: 63-82) samples tested positive for PCVs by quantitative PCR. In detail, 22 (27%, 95%CI: 18-37) were positive for PCV2, 58 (71%, 95%CI: 60-79) for PCV3, and 20 (24.4%, 95%CI 16-35) for both PCV2 and PCV3. On genome sequencing, different types and sub-types of PCV2 and PCV3 were identified, remarking a genetic diversity and hinting to a global circulation for the identified PCV strains. Overall, the high prevalence suggests that PCV2 and PCV3 infections are endemic in the wild boar population, posing risks for semi-intensive and free-range pig farming, typical of this region, due to contact with PCV-infected wild boar.
Keywords: Circoviridae; Southern Italy; porcine circoviruses; swine diseases; viral diseases; wild boar.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

