Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the "Calcification" Process
- PMID: 35454723
- PMCID: PMC9030872
- DOI: 10.3390/foods11081136
Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the "Calcification" Process
Abstract
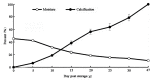
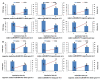
Chestnut "calcification" is the result of a series of physiological and biochemical changes during postharvest storage; however, the associated mechanisms are unclear. In this study, several potential calcification-related physicochemical parameters in chestnut, including moisture, cell wall materials, cellulose, lignin, and pectin, were measured. Transcriptome analysis was performed on chestnut seeds during different stages of storage. The results showed that the degree of calcification in the chestnut seeds was significantly negatively correlated with the moisture content (r = -0.961) at room temperature (20-25 °C) and a relative humidity of 50-60%. The accumulation of cell wall material in completely calcified seeds was 5.3 times higher than that of fresh seeds. The total content of cellulose and lignin increased during the storage process. Transcriptome analysis of 0% and 50% calcified chestnut was performed; a total of 1801 differentially expressed genes consisting of 805 up-regulated and 996 down-regulated genes were identified during the calcification process. Furthermore, response to water, water deprivation, and salt stress were most enriched by gene ontology (GO) and gene set enrichment analysis (GSEA). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to chestnut calcification included purine metabolism, RNA degradation, the mRNA surveillance pathway, starch and sucrose metabolism, arginine and proline metabolism, and fatty acid metabolism, and were detected. Most of the genes involved in cellulose synthase, lignin catabolism, and pectin catabolism were down-regulated, while only two important genes, scaffold11300 and scaffold0412, were up-regulated, which were annotated as cellulose and pectin synthase genes, respectively. These two genes may contribute to the increase of total cell wall material accumulation during chestnut calcification. The results provided new insights into chestnut calcification process and laid a foundation for further chestnut preservation.
Keywords: calcification; cellulose; chestnut; lignin; pectin; transcriptome.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Liu G.H., Fang J.Y. Spatial patterns of chestnut (Castanea millissima) and its species geographical distribution in China. Acta Ecol. Sin. 2001;21:164–170.
-
- Cui H., Li H., Wang Y., Li S., Xue C. Structural characterization and biological activity of galactoglucan from Castanea mollissima Blume. J. Carbohydr. Chem. 2019;38:398–411. doi: 10.1080/07328303.2019.1630838. - DOI
-
- Zhang Y., Yang Z., Liu G., Wu Y., Ouyang J. Inhibitory Effect of Chestnut (Castanea mollissima Blume) Inner Skin Extract on the Activity of α-Amylase, α-Glucosidase, Dipeptidyl Peptidase IV and In Vitro Digestibility of Starches. Food Chem. 2020;324:126847. doi: 10.1016/j.foodchem.2020.126847. - DOI - PubMed
-
- Wen X., Gu C., Zhu D., Liu P., Lai Y., Zeng Q. Water stress affects on cell membrane lipid oxidation and calcification of chestnut (Castanea mollissima Bl.) Postharvest Biol. Technol. 2017;126:34–39. doi: 10.1016/j.postharvbio.2016.12.005. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

