Deciphering Genetic Alterations of Hairy Cell Leukemia and Hairy Cell Leukemia-like Disorders in 98 Patients
- PMID: 35454811
- PMCID: PMC9028144
- DOI: 10.3390/cancers14081904
Deciphering Genetic Alterations of Hairy Cell Leukemia and Hairy Cell Leukemia-like Disorders in 98 Patients
Abstract
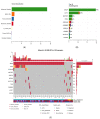
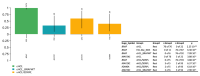
Hairy cell leukemia (cHCL) patients have, in most cases, a specific clinical and biological presentation with splenomegaly, anemia, leukopenia, neutropenia, monocytopenia and/or thrombocytopenia, identification of hairy cells that express CD103, CD123, CD25, CD11c and identification of the V600E mutation in the B-Raf proto-oncogene (BRAF) in 90% of cases. Monocytopenia is absent in vHCL and SDRPL patients and the abnormal cells do not express CD25 or CD123 and do not present the BRAFV600E mutation. Ten percent of cHCL patients are BRAFWT and the distinction between cHCL and HCL-like disorders including the variant form of HCL (vHCL) and splenic diffuse red pulp lymphoma (SDRPL) can be challenging. We performed deep sequencing in a large cohort of 84 cHCL and 16 HCL-like disorders to improve insights into the pathogenesis of the diseases. BRAF mutations were detected in 76/82 patients of cHCL (93%) and additional mutations were identified in Krüppel-like Factor 2 (KLF2) in 19 patients (23%) or CDKN1B in 6 patients (7.5%). Some KLF2 genetic alterations were localized on the cytidine deaminase (AID) consensus motif, suggesting AID-induced mutations. When analyzing sequential samples, a clonal evolution was identified in half of the cHCL patients (6/12 pts). Among the 16 patients with HCL-like disorders, we observed an enrichment of MAP2K1 mutations in vHCL/SDRPL (3/5 pts) and genes involved in the epigenetic regulation (KDM6A, EZH2, CREBBP, ARID1A) (3/5 pts). Furthermore, MAP2K1 mutations were associated with a bad prognosis and a shorter time to next treatment (TTNT) and progression-free survival (PFS), independently of the HCL classification.
Keywords: BRAF; HCL; KLF2; MAP2K1; genetic alterations; hairy cell leukemia; hairy cell leukemia variant (vHCL); hairy cell leukemia-like disorders; splenic diffuse red pulp lymphoma (SDRPL).
Conflict of interest statement
Xavier Troussard is a consultant for Innate Pharma, Astra Zeneca, Abbvie, the other authors declare that there are no conflicts of interest.
Figures




References
-
- Teras L.R., DeSantis C.E., Cerhan J.R., Morton L.M., Jemal A., Flowers C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes: 2016 US Lymphoid Malignancy Statistics by World Health Organization Subtypes. CA Cancer J. Clin. 2016;66:443–459. doi: 10.3322/caac.21357. - DOI - PubMed
-
- Troussard X. Epidemiology of Hairy Cell Leukemia (HCL) and HCL-Like Disorders. Med. Res. Arch. 2020;8:1–11. doi: 10.18103/mra.v8i10.2259. - DOI
-
- Matutes E., Morilla R., Owusu-Ankomah K., Houliham H., Meeus P., Catovsky D. The immunophenotype of hairy cell leukemia (HCL). Proposal for a scoring system to distinguish HCL from B-cell disorders with hairy or villous lymphocytes. Leuk. Lymphoma. 1994;14:57–61. - PubMed
-
- Forconi F., Sozzi E., Cencini E., Zaja F., Intermesoli T., Stelitano C., Rigacci L., Gherlinzoni F., Cantaffa R., Baraldi A., et al. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood. 2009;114:4696–4702. doi: 10.1182/blood-2009-03-212449. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

