Application of Proteogenomics to Urine Analysis towards the Identification of Novel Biomarkers of Prostate Cancer: An Exploratory Study
- PMID: 35454907
- PMCID: PMC9031064
- DOI: 10.3390/cancers14082001
Application of Proteogenomics to Urine Analysis towards the Identification of Novel Biomarkers of Prostate Cancer: An Exploratory Study
Abstract
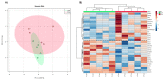
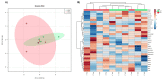
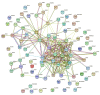
To identify new protein targets for PCa detection, first, a shotgun discovery experiment was performed to characterize the urinary proteome of PCa patients. This revealed 18 differentially abundant urinary proteins in PCa patients. Second, selected targets were clinically tested by immunoblot, and the soluble E-cadherin fragment was detected for the first time in the urine of PCa patients. Third, the proteogenome landscape of these PCa patients was characterized, revealing 1665 mutant protein isoforms. Statistical analysis revealed 6 differentially abundant mutant protein isoforms in PCa patients. Analysis of the likely effects of mutations on protein function and PPIs involving the dysregulated mutant protein isoforms suggests a protective role of mutations HSPG2*Q1062H and VASN*R161Q and an adverse role of AMBP*A286G and CD55*S162L in PCa patients. This work originally characterized the urinary proteome, focusing on the proteogenome profile of PCa patients, which is usually overlooked in the analysis of PCa and body fluids. Combined analysis of mass spectrometry data using two different software packages was performed for the first time in the context of PCa, which increased the robustness of the data analysis. The application of proteogenomics to urine proteomic analysis can be very enriching in mutation-related diseases such as cancer.
Keywords: biomarker; human; immunoblot; label-free quantitation; prostate cancer; proteogenome; proteome; urine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
- SFRH/BD/136904/2018/Fundação para a Ciência e Tecnologia
- UIDP/00051/2020/Fundação para a Ciência e Tecnologia
- IF/00286/2015/Fundação para a Ciência e Tecnologia
- NORTE-01-0145-FEDER-000003/Norte2020 - Norte2020-Projetos Estruturados de I&D&I
- UIDB/50006/2020/Fundação para a Ciência e Tecnologia
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

