RNA World Modeling: A Comparison of Two Complementary Approaches
- PMID: 35455198
- PMCID: PMC9027272
- DOI: 10.3390/e24040536
RNA World Modeling: A Comparison of Two Complementary Approaches
Abstract
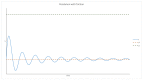
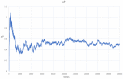
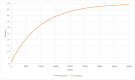
The origin of life remains one of the major scientific questions in modern biology. Among many hypotheses aiming to explain how life on Earth started, RNA world is probably the most extensively studied. It assumes that, in the very beginning, RNA molecules served as both enzymes and as genetic information carriers. However, even if this is true, there are many questions that still need to be answered-for example, whether the population of such molecules could achieve stability and retain genetic information for many generations, which is necessary in order for evolution to start. In this paper, we try to answer this question based on the parasite-replicase model (RP model), which divides RNA molecules into enzymes (RNA replicases) capable of catalyzing replication and parasites that do not possess replicase activity but can be replicated by RNA replicases. We describe the aforementioned system using partial differential equations and, based on the analysis of the simulation, surmise general rules governing its evolution. We also compare this approach with one where the RP system is modeled and implemented using a multi-agent modeling technique. We show that approaching the description and analysis of the RP system from different perspectives (microscopic represented by MAS and macroscopic depicted by PDE) provides consistent results. Therefore, applying MAS does not lead to erroneous results and allows us to study more complex situations where many cases are concerned, which would not be possible through the PDE model.
Keywords: RNA world; multi-agent systems; partial differential equations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
Simulating the origins of life: The dual role of RNA replicases as an obstacle to evolution.PLoS One. 2017 Jul 10;12(7):e0180827. doi: 10.1371/journal.pone.0180827. eCollection 2017. PLoS One. 2017. PMID: 28700697 Free PMC article.
-
Multi-agent approach to sequence structure simulation in the RNA World hypothesis.PLoS One. 2020 Aug 28;15(8):e0238253. doi: 10.1371/journal.pone.0238253. eCollection 2020. PLoS One. 2020. PMID: 32857812 Free PMC article.
-
Monte Carlo simulation of early molecular evolution in the RNA World.Biosystems. 2007 Jul-Aug;90(1):28-39. doi: 10.1016/j.biosystems.2006.06.005. Epub 2006 Jun 23. Biosystems. 2007. PMID: 17014951
-
[Unsolved Puzzles of Qβ Replicase].Mol Biol (Mosk). 2019 Nov-Dec;53(6):899-910. doi: 10.1134/S0026898419060041. Mol Biol (Mosk). 2019. PMID: 31876271 Review. Russian.
-
In search of an RNA replicase ribozyme.Chem Biol. 2003 Jan;10(1):5-14. doi: 10.1016/s1074-5521(03)00003-6. Chem Biol. 2003. PMID: 12573693 Review.
Cited by
-
RNA World with Inhibitors.Entropy (Basel). 2024 Nov 23;26(12):1012. doi: 10.3390/e26121012. Entropy (Basel). 2024. PMID: 39766641 Free PMC article.
-
Advancements in bioinformatics and computational biology: 15th annual Polish Bioinformatic Society Symposium.Bioinform Adv. 2024 Jan 13;4(1):vbad187. doi: 10.1093/bioadv/vbad187. eCollection 2024. Bioinform Adv. 2024. PMID: 38239846 Free PMC article.
References
-
- Szostak N., Wasik S., Blazewicz J. Understanding Life: A Bioinformatics Perspective. Eur. Rev. 2017;25:231–245. doi: 10.1017/S1062798716000570. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

