CFTR Modulation Reduces SARS-CoV-2 Infection in Human Bronchial Epithelial Cells
- PMID: 35456026
- PMCID: PMC9028056
- DOI: 10.3390/cells11081347
CFTR Modulation Reduces SARS-CoV-2 Infection in Human Bronchial Epithelial Cells
Abstract
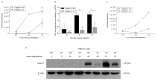
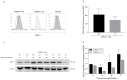
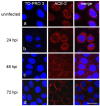
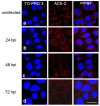
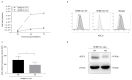
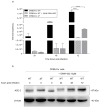
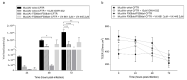
People with cystic fibrosis should be considered at increased risk of developing severe symptoms of COVID-19. Strikingly, a broad array of evidence shows reduced spread of SARS-CoV-2 in these subjects, suggesting a potential role for CFTR in the regulation of SARS-CoV-2 infection/replication. Here, we analyzed SARS-CoV-2 replication in wild-type and CFTR-modified human bronchial epithelial cell lines and primary cells to investigate SARS-CoV-2 infection in people with cystic fibrosis. Both immortalized and primary human bronchial epithelial cells expressing wt or F508del-CFTR along with CRISPR/Cas9 CFTR-ablated clones were infected with SARS-CoV-2 and samples were harvested before and from 24 to 72 h post-infection. CFTR function was also inhibited in wt-CFTR cells with the CFTR-specific inhibitor IOWH-032 and partially restored in F508del-CFTR cells with a combination of CFTR modulators (VX-661+VX-445). Viral load was evaluated by real-time RT-PCR in both supernatant and cell extracts, and ACE-2 expression was analyzed by both western blotting and flow cytometry. SARS-CoV-2 replication was reduced in CFTR-modified bronchial cells compared with wild-type cell lines. No major difference in ACE-2 expression was detected before infection between wild-type and CFTR-modified cells, while a higher expression in wild-type compared to CFTR-modified cells was detectable at 72 h post-infection. Furthermore, inhibition of CFTR channel function elicited significant inhibition of viral replication in cells with wt-CFTR, and correction of CFTR function in F508del-CFTR cells increased the release of SARS-CoV-2 viral particles. Our study provides evidence that CFTR expression/function is involved in the regulation of SARS-CoV-2 replication, thus providing novel insights into the role of CFTR in SARS-CoV-2 infection and the development of therapeutic strategies for COVID-19.
Keywords: ACE-2; CFTR; CFTR inhibitor; SARS-CoV-2 virus; cystic fibrosis; human bronchial epithelial cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hui D.S., Ahzar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
-
- W.H.O WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. 2020. [(accessed on 11 March 2020)]. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re....
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

