Pangenomics in Microbial and Crop Research: Progress, Applications, and Perspectives
- PMID: 35456404
- PMCID: PMC9031676
- DOI: 10.3390/genes13040598
Pangenomics in Microbial and Crop Research: Progress, Applications, and Perspectives
Abstract
Advances in sequencing technologies and bioinformatics tools have fueled a renewed interest in whole genome sequencing efforts in many organisms. The growing availability of multiple genome sequences has advanced our understanding of the within-species diversity, in the form of a pangenome. Pangenomics has opened new avenues for future research such as allowing dissection of complex molecular mechanisms and increased confidence in genome mapping. To comprehensively capture the genetic diversity for improving plant performance, the pangenome concept is further extended from species to genus level by the inclusion of wild species, constituting a super-pangenome. Characterization of pangenome has implications for both basic and applied research. The concept of pangenome has transformed the way biological questions are addressed. From understanding evolution and adaptation to elucidating host-pathogen interactions, finding novel genes or breeding targets to aid crop improvement to design effective vaccines for human prophylaxis, the increasing availability of the pangenome has revolutionized several aspects of biological research. The future availability of high-resolution pangenomes based on reference-level near-complete genome assemblies would greatly improve our ability to address complex biological problems.
Keywords: NGS; biological research; evolution; genome sequence; germplasm; novel genes; pangenome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
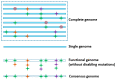
 represents a disabling mutation that disrupts the gene function.
represents a disabling mutation that disrupts the gene function.  ,
,  , and
, and  depict various sequence polymorphisms.
depict various sequence polymorphisms.
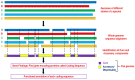
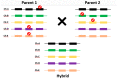
 represents “functional block” leading to null or altered protein function. In a real scenario, accumulation of a similar effect with many gene families leads to reduced vigor in inbreeds and heterosis in hybrid. Pangenomics can help to unravel heterosis in a phenotypic trait by discovering new gene variants.
represents “functional block” leading to null or altered protein function. In a real scenario, accumulation of a similar effect with many gene families leads to reduced vigor in inbreeds and heterosis in hybrid. Pangenomics can help to unravel heterosis in a phenotypic trait by discovering new gene variants.References
-
- Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus Agalactiae: Implications for the Microbial “pangenome”. Proc. Natl. Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

