Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum
- PMID: 35456413
- PMCID: PMC9028111
- DOI: 10.3390/genes13040607
Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum
Abstract
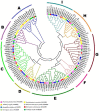
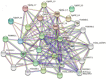
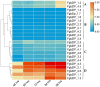
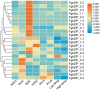
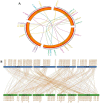
The basic leucine zipper (bZIP) is a widely found transcription factor family that plays regulatory roles in a variety of cellular processes including cell growth and development and various stress responses. However, the bZIP gene family has not been well studied at a genome-wide scale in Fusarium graminearum (Fg), a potent pathogen of cereal grains. In the present study, we conducted a genome-wide identification, characterization, and expression profiling of 22 F. graminearum bZIP (FgbZIP) genes at different developmental stages and under various abiotic stresses. All identified FgbZIPs were categorized into nine groups based on their sequence similarity and phylogenetic tree analysis. Furthermore, the gene structure analysis, conserved motif analysis, chromosomal localization, protein network studies, and synteny analysis were performed. The symmetry of the exon and intron varied with the phylogenetic groups. The post-translational modifications (PTMs) analysis also predicted several phosphorylation sites in FgbZIPs, indicating their functional diversity in cellular processes. The evolutionary study identified many orthogroups among eight species and also predicted several gene duplication events in F. graminearum. The protein modeling indicated the presence of a higher number of α-helices and random coils in their structures. The expression patterns of FgbZIP genes showed that 5 FgbZIP genes, including FgbZIP_1.1, FgbZIP_1.3, FgbZIP_2.6 FgbZIP_3.1 and FgbZIP_4.3, had high expression at different growth and conidiogenesis stages. Similarly, eight genes including FgbZIP_1.1, FgbZIP_1.6, FgbZIP_2.3, FgbZIP_2.4, FgbZIP_4.1, FgbZIP_4.2, FgbZIP_4.3 and FgbZIP_4.6 demonstrated their putative role in response to various abiotic stresses. In summary, these results provided basic information regarding FgbZIPs which are helpful for further functional analysis.
Keywords: Fusarium graminearum; abiotic stress; bZIP; expression analysis; phylogenetic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera).BMC Genomics. 2014 Apr 13;15:281. doi: 10.1186/1471-2164-15-281. BMC Genomics. 2014. PMID: 24725365 Free PMC article.
-
Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear (Pyrus bretschneideri).BMC Plant Biol. 2021 Sep 9;21(1):413. doi: 10.1186/s12870-021-03191-3. BMC Plant Biol. 2021. PMID: 34503442 Free PMC article.
-
Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes.BMC Genomics. 2015 Dec 10;16:1053. doi: 10.1186/s12864-015-2258-x. BMC Genomics. 2015. PMID: 26651343 Free PMC article.
-
Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress.Protoplasma. 2017 Jan;254(1):3-16. doi: 10.1007/s00709-015-0920-4. Epub 2015 Dec 15. Protoplasma. 2017. PMID: 26669319 Review.
-
Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function.Biochim Biophys Acta. 2016 Jan;1860(1 Pt A):46-56. doi: 10.1016/j.bbagen.2015.10.014. Epub 2015 Oct 20. Biochim Biophys Acta. 2016. PMID: 26493723 Review.
Cited by
-
Genome-Wide Profiling of bZIP Transcription Factors and FocbZIP11's Impact on Fusarium TR4 Pathogenicity.Int J Mol Sci. 2025 Feb 9;26(4):1452. doi: 10.3390/ijms26041452. Int J Mol Sci. 2025. PMID: 40003918 Free PMC article.
-
Identifying transcription factors associated with Fusarium graminearum virus 2 accumulation in Fusarium graminearum by phenome-based investigation.Virus Res. 2023 Mar;326:199061. doi: 10.1016/j.virusres.2023.199061. Epub 2023 Feb 8. Virus Res. 2023. PMID: 36738934 Free PMC article.
-
Exploring Aluminum Tolerance Mechanisms in Plants with Reference to Rice and Arabidopsis: A Comprehensive Review of Genetic, Metabolic, and Physiological Adaptations in Acidic Soils.Plants (Basel). 2024 Jun 25;13(13):1760. doi: 10.3390/plants13131760. Plants (Basel). 2024. PMID: 38999600 Free PMC article. Review.
-
In-Depth Characterization of bZIP Genes in the Context of Endoplasmic Reticulum (ER) Stress in Brassica campestris ssp. chinensis.Plants (Basel). 2024 Apr 22;13(8):1160. doi: 10.3390/plants13081160. Plants (Basel). 2024. PMID: 38674568 Free PMC article.
-
Genome-Wide Identification and Functional Analysis of the bZIP Transcription Factor Family in Rice Bakanae Disease Pathogen, Fusarium fujikuroi.Int J Mol Sci. 2022 Jun 15;23(12):6658. doi: 10.3390/ijms23126658. Int J Mol Sci. 2022. PMID: 35743103 Free PMC article.
References
-
- Rauwane M.E., Ogugua U.V., Kalu C.M., Ledwaba L.K., Woldesemayat A.A., Ntushelo K. Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms. 2020;8:305. doi: 10.3390/microorganisms8020305. - DOI - PMC - PubMed
-
- Dweba C., Figlan S., Shimelis H., Motaung T., Sydenham S., Mwadzingeni L., Tsilo T. Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 2017;91:114–122. doi: 10.1016/j.cropro.2016.10.002. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

