Transcriptional Profiling and Deriving a Seven-Gene Signature That Discriminates Active and Latent Tuberculosis: An Integrative Bioinformatics Approach
- PMID: 35456421
- PMCID: PMC9032611
- DOI: 10.3390/genes13040616
Transcriptional Profiling and Deriving a Seven-Gene Signature That Discriminates Active and Latent Tuberculosis: An Integrative Bioinformatics Approach
Abstract
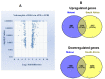
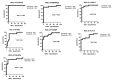
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (M.tb.). Our integrative analysis aims to identify the transcriptional profiling and gene expression signature that distinguish individuals with active TB (ATB) disease, and those with latent tuberculosis infection (LTBI). In the present study, we reanalyzed a microarray dataset (GSE37250) from GEO database and explored the data for differential gene expression analysis between those with ATB and LTBI derived from Malawi and South African cohorts. We used BRB array tool to distinguish DEGs (differentially expressed genes) between ATB and LTBI. Pathway enrichment analysis of DEGs was performed using DAVID bioinformatics tool. The protein-protein interaction (PPI) network of most upregulated genes was constructed using STRING analysis. We have identified 375 upregulated genes and 152 downregulated genes differentially expressed between ATB and LTBI samples commonly shared among Malawi and South African cohorts. The constructed PPI network was significantly enriched with 76 nodes connected to 151 edges. The enriched GO term/pathways were mainly related to expression of IFN stimulated genes, interleukin-1 production, and NOD-like receptor signaling pathway. Downregulated genes were significantly enriched in the Wnt signaling, B cell development, and B cell receptor signaling pathways. The short-listed DEGs were validated in a microarray data from an independent cohort (GSE19491). ROC curve analysis was done to assess the diagnostic accuracy of the gene signature in discrimination of active and latent tuberculosis. Thus, we have derived a seven-gene signature, which included five upregulated genes FCGR1B, ANKRD22, CARD17, IFITM3, TNFAIP6 and two downregulated genes FCGBP and KLF12, as a biomarker for discrimination of active and latent tuberculosis. The identified genes have a sensitivity of 80-100% and specificity of 80-95%. Area under the curve (AUC) value of the genes ranged from 0.84 to 1. This seven-gene signature has a high diagnostic accuracy in discrimination of active and latent tuberculosis.
Keywords: active TB; bioinformatics; biomarkers; differentially expressed genes; latent TB infection; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Health Organization . Global Tuberculosis Report 2021. World Health Organization; Geneva, Switzerland: 2021. [(accessed on 1 January 2022)]. Available online: https://www.who.int/publications/i/item/9789240037021.
-
- World Health Organization . Global Tuberculosis Report 2018. World Health Organization; Geneva, Switzerland: 2018. [(accessed on 31 May 2021)]. Available online: https://apps.who.int/iris/handle/10665/274453.
-
- Drain P.K., Bajema K.L., Dowdy D., Dheda K., Naidoo K., Schumacher S.G., Ma S., Meermeier E., Lewinsohn D.M., Sherman D.R. Incipient and subclinical tuberculosis: A clinical review of early stages and progression of infection. Clin. Microbiol. Rev. 2018;31:e00021-18. doi: 10.1128/CMR.00021-18. - DOI - PMC - PubMed
-
- Kurt T., Frieden T.R., World Health Organization . In: Toman’s Tuberculosis: Case Detection, Treatment, and Monitoring: Questions and Answers. 2nd ed. Frieden T., editor. World Health Organization; Geneva, Switzerland: 2004.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

