Integrated Analysis of the ceRNA Network and M-7474 Function in Testosterone-Mediated Fat Deposition in Pigs
- PMID: 35456474
- PMCID: PMC9032878
- DOI: 10.3390/genes13040668
Integrated Analysis of the ceRNA Network and M-7474 Function in Testosterone-Mediated Fat Deposition in Pigs
Abstract
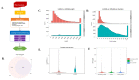
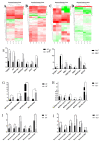
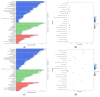
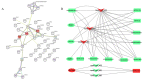
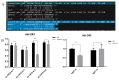
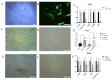
Castration can significantly enhance fat deposition in pigs, and the molecular mechanism of fat deposition caused by castration and its influence on fat deposition in different parts of pigs remain unclear. RNA-seq was performed on adipose tissue from different parts of castrated and intact Yorkshire pigs. Different ceRNA networks were constructed for different fat parts. GO and KEGG pathway annotations suggested that testosterone elevates cell migration and affects differentiation and apoptosis in back fat, while it predisposes animals to glycolipid metabolism disorders and increases the expression of inflammatory cytokines in abdominal fat. The interaction between M-7474, novel_miR_243 and SGK1 was verified by dual fluorescence experiments. This ceRNA relationship has also been demonstrated in porcine preadipocytes. Overexpression of M-7474 significantly inhibited the differentiation of preadipocytes compared to the control group. When 100 nM testosterone was added during preadipocyte differentiation, the expression of M-7474 was increased, and preadipocyte differentiation was significantly inhibited. Testosterone can affect preadipocyte differentiation by upregulating the expression of M-7474, sponging novel-miR-243, and regulating the expression of genes such as SGK1. At the same time, HSD11B1 and SLC2A4 may also be regulated by the corresponding lncRNA and miRNA, which ultimately affects glucose uptake by adipocytes and leads to obesity.
Keywords: castration; ceRNA; fat deposition; lncRNA; testosterone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Robison O.W., Johnson B., Lubritz D. Genetic parameters for testosterone production in boars. J. Anim. Sci. 1991;69:3220–3224. - PubMed
-
- Yeh S., Tsai M.Y., Xu Q., Mu X.M., Lardy H., Huang K.E., Lin H., Yeh S.D., Altuwaijri S., Zhou X., et al. Generation and characterization of androgen receptor knockout (ARKO) mice: An in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci. USA. 2002;99:13498–13503. doi: 10.1073/pnas.212474399. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

