m5CRegpred: Epitranscriptome Target Prediction of 5-Methylcytosine (m5C) Regulators Based on Sequencing Features
- PMID: 35456483
- PMCID: PMC9025882
- DOI: 10.3390/genes13040677
m5CRegpred: Epitranscriptome Target Prediction of 5-Methylcytosine (m5C) Regulators Based on Sequencing Features
Abstract
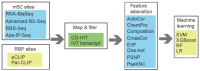
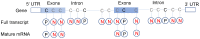
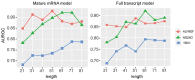
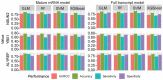
5-methylcytosine (m5C) is a common post-transcriptional modification observed in a variety of RNAs. m5C has been demonstrated to be important in a variety of biological processes, including RNA structural stability and metabolism. Driven by the importance of m5C modification, many projects focused on the m5C sites prediction were reported before. To better understand the upstream and downstream regulation of m5C, we present a bioinformatics framework, m5CRegpred, to predict the substrate of m5C writer NSUN2 and m5C readers YBX1 and ALYREF for the first time. After features comparison, window lengths selection and algorism comparison on the mature mRNA model, our model achieved AUROC scores 0.869, 0.724 and 0.889 for NSUN2, YBX1 and ALYREF, respectively in an independent test. Our work suggests the substrate of m5C regulators can be distinguished and may help the research of m5C regulators in a special condition, such as substrates prediction of hyper- or hypo-expressed m5C regulators in human disease.
Keywords: 5-methylcytosine; machine learning; readers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Heissenberger C., Liendl L., Nagelreiter F., Gonskikh Y., Yang G., Stelzer E.M., Krammer T.L., Micutkova L., Vogt S., Kreil D.P., et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019;47:11807–11825. doi: 10.1093/nar/gkz1043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

