Methodologies for the De novo Discovery of Transposable Element Families
- PMID: 35456515
- PMCID: PMC9025800
- DOI: 10.3390/genes13040709
Methodologies for the De novo Discovery of Transposable Element Families
Abstract
The discovery and characterization of transposable element (TE) families are crucial tasks in the process of genome annotation. Careful curation of TE libraries for each organism is necessary as each has been exposed to a unique and often complex set of TE families. De novo methods have been developed; however, a fully automated and accurate approach to the development of complete libraries remains elusive. In this review, we cover established methods and recent developments in de novo TE analysis. We also present various methodologies used to assess these tools and discuss opportunities for further advancement of the field.
Keywords: curation; de novo methods; genome annotation; repeats; signature-based methods; transposable element; transposon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

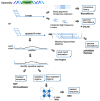


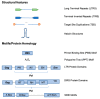

References
-
- International Barley Genome Sequencing Consortium. Mayer K.F., Waugh R., Brown J.W., Schulman A., Langridge P., Platzer M., Fincher G.B., Muehlbauer G.J., Sato K., et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

