eGFP Gene Integration in HO: A Metabolomic Impact?
- PMID: 35456831
- PMCID: PMC9032140
- DOI: 10.3390/microorganisms10040781
eGFP Gene Integration in HO: A Metabolomic Impact?
Abstract
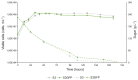
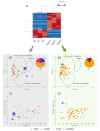
Integrating fluorescent genes including eGFP in the yeast genome is common practice for various applications, including cell visualization and population monitoring. The transformation of a commercial S. cerevisiae strain by integrating a cassette including a gene encoding an EGFP protein in the HO gene was carried out using CRISPR-Cas9 technology. Although this type of integration is often used and described as neutral at the phenotypic level of the cell, we have highlighted that under alcoholic fermentation (in a Chardonnay must), it has an impact on the exometabolome. We observed 41 and 82 unique biomarkers for the S3 and S3GFP strains, respectively, as well as 28 biomarkers whose concentrations varied significantly between the wild-type and the modified strains. These biomarkers were mainly found to correspond to peptides. Despite similar phenotypic growth and fermentation parameters, high-resolution mass spectrometry allowed us to demonstrate, for the first time, that the peptidome is modified when integrating this cassette in the HO gene.
Keywords: CRISPR-Cas9 technology; HO gene; S. cerevisiae; eGFP; metabolomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lorang J.M., Tuori R.P., Martinez J.P., Sawyer T.L., Redman R.S., Rollins J.A., Wolpert T.J., Johnson K.B., Rodriguez R.J., Dickman M.B., et al. Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 2001;67:1987–1994. doi: 10.1128/AEM.67.5.1987-1994.2001. - DOI - PMC - PubMed
-
- Tatchell K., Robinson L.C. Methods in Enzymology. Volume 351. Elsevier; Amsterdam, The Netherlands: 2002. Use of green fluorescent protein in living yeast cells; pp. 661–683. - PubMed
-
- Petitgonnet C., Klein G.L., Roullier-Gall C., Schmitt-Kopplin P., Quintanilla-Casas B., Vichi S., Julien-David D., Alexandre H. Influence of cell-cell contact between L. thermotolerans and S. cerevisiae on yeast interactions and the exo-metabolome. Food Microbiol. 2019;83:122–133. doi: 10.1016/j.fm.2019.05.005. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

