Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds
- PMID: 35456898
- PMCID: PMC9028948
- DOI: 10.3390/ijms23084081
Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds
Abstract
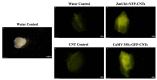
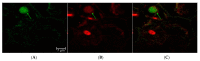
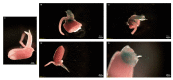
CRISPR-Cas gene editing technologies offer the potential to modify crops precisely; however, in vitro plant transformation and regeneration techniques present a bottleneck due to the lengthy and genotype-specific tissue culture process. Ideally, in planta transformation can bypass tissue culture and directly lead to transformed plants, but efficient in planta delivery and transformation remains a challenge. This study investigates transformation methods that have the potential to directly alter germline cells, eliminating the challenge of in vitro plant regeneration. Recent studies have demonstrated that carbon nanotubes (CNTs) loaded with plasmid DNA can diffuse through plant cell walls, facilitating transient expression of foreign genetic elements in plant tissues. To test if this approach is a viable technique for in planta transformation, CNT-mediated plasmid DNA delivery into rice tissues was performed using leaf and excised-embryo infiltration with reporter genes. Quantitative and qualitative data indicate that CNTs facilitate plasmid DNA delivery in rice leaf and embryo tissues, resulting in transient GFP, YFP, and GUS expression. Experiments were also initiated with CRISPR-Cas vectors targeting the phytoene desaturase (PDS) gene for CNT delivery into mature embryos to create heritable genetic edits. Overall, the results suggest that CNT-based delivery of plasmid DNA appears promising for in planta transformation, and further optimization can enable high-throughput gene editing to accelerate functional genomics and crop improvement activities.
Keywords: CRISPR/Cas9; carbon nanotubes (CNTs); gene editing; phytoene desaturase (PDS); rice (Oryza sativa).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- FAO . The Future of Food and Agriculture: Trends and Challenges. Volume 296. FAO; Rome, Italy: 2017. pp. 1–180.
-
- United Nations World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. 2015. [(accessed on 5 December 2020)]. Available online: https://population.un.org/wpp/Publications/Files/Key_Findings_WPP_2015.pdf.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

