Small RNAs beyond Model Organisms: Have We Only Scratched the Surface?
- PMID: 35457265
- PMCID: PMC9029176
- DOI: 10.3390/ijms23084448
Small RNAs beyond Model Organisms: Have We Only Scratched the Surface?
Abstract
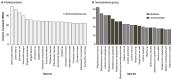
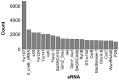
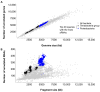
Small RNAs (sRNAs) are essential regulators in the adaptation of bacteria to environmental changes and act by binding targeted mRNAs through base complementarity. Approximately 550 distinct families of sRNAs have been identified since their initial characterization in the 1980s, accelerated by the emergence of RNA-sequencing. Small RNAs are found in a wide range of bacterial phyla, but they are more prominent in highly researched model organisms compared to the rest of the sequenced bacteria. Indeed, Escherichia coli and Salmonella enterica contain the highest number of sRNAs, with 98 and 118, respectively, with Enterobacteriaceae encoding 145 distinct sRNAs, while other bacteria families have only seven sRNAs on average. Although the past years brought major advances in research on sRNAs, we have perhaps only scratched the surface, even more so considering RNA annotations trail behind gene annotations. A distinctive trend can be observed for genes, whereby their number increases with genome size, but this is not observable for RNAs, although they would be expected to follow the same trend. In this perspective, we aimed at establishing a more accurate representation of the occurrence of sRNAs in bacteria, emphasizing the potential for novel sRNA discoveries.
Keywords: genetic regulation; non-coding RNA; small RNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
APERO: a genome-wide approach for identifying bacterial small RNAs from RNA-Seq data.Nucleic Acids Res. 2019 Sep 5;47(15):e88. doi: 10.1093/nar/gkz485. Nucleic Acids Res. 2019. PMID: 31147705 Free PMC article.
-
A Modular Genetic System for High-Throughput Profiling and Engineering of Multi-Target Small RNAs.Methods Mol Biol. 2018;1737:373-391. doi: 10.1007/978-1-4939-7634-8_21. Methods Mol Biol. 2018. PMID: 29484604
-
Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium.RNA Biol. 2016;13(3):331-42. doi: 10.1080/15476286.2016.1144010. Epub 2016 Feb 6. RNA Biol. 2016. PMID: 26853797 Free PMC article.
-
Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica.EcoSal Plus. 2020 Mar;9(1):10.1128/ecosalplus.ESP-0030-2019. doi: 10.1128/ecosalplus.ESP-0030-2019. EcoSal Plus. 2020. PMID: 32213244 Free PMC article. Review.
-
Prevalence of small base-pairing RNAs derived from diverse genomic loci.Biochim Biophys Acta Gene Regul Mech. 2020 Jul;1863(7):194524. doi: 10.1016/j.bbagrm.2020.194524. Epub 2020 Mar 5. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32147527 Free PMC article. Review.
Cited by
-
Genomic basis of environmental adaptation in the widespread poly-extremophilic Exiguobacterium group.ISME J. 2024 Jan 8;18(1):wrad020. doi: 10.1093/ismejo/wrad020. ISME J. 2024. PMID: 38365240 Free PMC article.
-
High-Resolution Small RNAs Landscape Provides Insights into Alkane Adaptation in the Marine Alkane-Degrader Alcanivorax dieselolei B-5.Int J Mol Sci. 2022 Dec 15;23(24):15995. doi: 10.3390/ijms232415995. Int J Mol Sci. 2022. PMID: 36555635 Free PMC article.
-
Battle for Metals: Regulatory RNAs at the Front Line.Front Cell Infect Microbiol. 2022 Jul 5;12:952948. doi: 10.3389/fcimb.2022.952948. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35865816 Free PMC article. Review.
-
Developing New Tools to Fight Human Pathogens: A Journey through the Advances in RNA Technologies.Microorganisms. 2022 Nov 21;10(11):2303. doi: 10.3390/microorganisms10112303. Microorganisms. 2022. PMID: 36422373 Free PMC article. Review.
-
MicroRNA gatekeepers: Orchestrating rhizospheric dynamics.J Integr Plant Biol. 2025 Mar;67(3):845-876. doi: 10.1111/jipb.13860. Epub 2025 Feb 21. J Integr Plant Biol. 2025. PMID: 39981727 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

