New Insights into the Structure and Assembly of Bacteriophage P1
- PMID: 35458408
- PMCID: PMC9024508
- DOI: 10.3390/v14040678
New Insights into the Structure and Assembly of Bacteriophage P1
Abstract
Bacteriophage P1 is the premier transducing phage of E. coli. Despite its prominence in advancing E. coli genetics, modern molecular techniques have not been applied to thoroughly understand P1 structure. Here, we report the proteome of the P1 virion as determined by liquid chromatography tandem mass-spectrometry. Additionally, a library of single-gene knockouts identified the following five previously unknown essential genes: pmgA, pmgB, pmgC, pmgG, and pmgR. In addition, proteolytic processing of the major capsid protein is a known feature of P1 morphogenesis, and we identified the processing site by N-terminal sequencing to be between E120 and S121, producing a 448-residue, 49.3 kDa mature peptide. Furthermore, the P1 defense against restriction (Dar) system consists of six known proteins that are incorporated into the virion during morphogenesis. The largest of these, DarB, is a 250 kDa protein that is believed to translocate into the cell during infection. DarB deletions indicated the presence of an N-terminal packaging signal, and the N-terminal 30 residues of DarB are shown to be sufficient for directing a heterologous reporter protein to the capsid. Taken together, the data expand on essential structural P1 proteins as well as introduces P1 as a nanomachine for cellular delivery.
Keywords: bacteriophage P1; bacteriophage structure; capsid targeting sequence; defense against restriction; proteomics; transmission electron microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



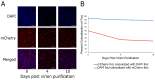
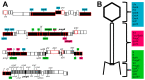
References
-
- Yarmolinsky M.B., Sternberg N. Bacteriophage P1. In: Calendar R., editor. The Bacteriophages. Springer; Boston, MA, USA: 1988. pp. 291–438.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

