Emergence of Two Distinct SARS-CoV-2 Gamma Variants and the Rapid Spread of P.1-like-II SARS-CoV-2 during the Second Wave of COVID-19 in Santa Catarina, Southern Brazil
- PMID: 35458424
- PMCID: PMC9029728
- DOI: 10.3390/v14040695
Emergence of Two Distinct SARS-CoV-2 Gamma Variants and the Rapid Spread of P.1-like-II SARS-CoV-2 during the Second Wave of COVID-19 in Santa Catarina, Southern Brazil
Abstract
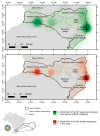
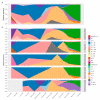
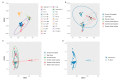
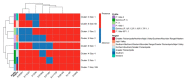
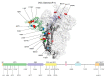
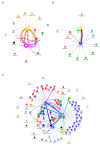
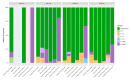
The western mesoregion of the state of Santa Catarina (SC), Southern Brazil, was heavily affected as a whole by the COVID-19 pandemic in early 2021. This study aimed to evaluate the dynamics of the SARS-CoV-2 virus spreading patterns in the SC state from March 2020 to April 2021 using genomic surveillance. During this period, there were 23 distinct variants, including Beta and Gamma, among which the Gamma and related lineages were predominant in the second pandemic wave within SC. A regionalization of P.1-like-II in the Western SC region was observed, concomitant to the increase in cases, mortality, and the case fatality rate (CFR) index. This is the first evidence of the regionalization of the SARS-CoV-2 transmission in SC and it highlights the importance of tracking the variants, dispersion, and impact of SARS-CoV-2 on the public health systems.
Keywords: SARS-CoV-2; SARS-CoV-2 P.1 variant; genome; surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Health Organization WHO Coronavirus (COVID-19) [(accessed on 12 December 2021)]. Available online: https://www.who.int.
-
- National Council of Health Secretaries-Covid-19 CONASS Panel. [(accessed on 4 December 2021)]. Available online: https://www.conass.org.br/painelconasscovid19/
-
- Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of healthcare associated COVID-19: A prospective genomic surveillance study. Lancet Infect. Dis. 2021;21:e36. Corrected and republished from Lancet Infect Dis. 2020, 20, 1263–1272. - PMC - PubMed
-
- Global Initiative on Sharing Avian Influenza Data. 2008. [(accessed on 27 November 2021)]. Available online: https://www.gisaid.org.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

