Early Emergence Phase of SARS-CoV-2 Delta Variant in Florida, US
- PMID: 35458495
- PMCID: PMC9028683
- DOI: 10.3390/v14040766
Early Emergence Phase of SARS-CoV-2 Delta Variant in Florida, US
Abstract
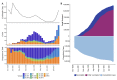
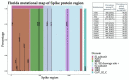
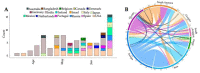
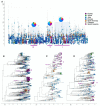
SARS-CoV-2, the causative agent of COVID-19, emerged in late 2019. The highly contagious B.1.617.2 (Delta) variant of concern (VOC) was first identified in October 2020 in India and subsequently disseminated worldwide, later becoming the dominant lineage in the US. Understanding the local transmission dynamics of early SARS-CoV-2 introductions may inform actionable mitigation efforts during subsequent pandemic waves. Yet, despite considerable genomic analysis of SARS-CoV-2 in the US, several gaps remain. Here, we explore the early emergence of the Delta variant in Florida, US using phylogenetic analysis of representative Florida and globally sampled genomes. We find multiple independent introductions into Florida primarily from North America and Europe, with a minority originating from Asia. These introductions led to three distinct clades that demonstrated varying relative rates of transmission and possessed five distinct substitutions that were 3-21 times more prevalent in the Florida sample as compared to the global sample. Our results underscore the benefits of routine viral genomic surveillance to monitor epidemic spread and support the need for more comprehensive genomic epidemiology studies of emerging variants. In addition, we provide a model of epidemic spread of newly emerging VOCs that can inform future public health responses.
Keywords: Delta; Florida; SARS-CoV-2; early emergence; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Johns Hopkins COVID-19 Map. Johns Hopkins Coronavirus Resource Center. [(accessed on 26 July 2021)]. Available online: https://coronavirus.jhu.edu/
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

