Longitudinal Survey of Coronavirus Circulation and Diversity in Insectivorous Bat Colonies in Zimbabwe
- PMID: 35458511
- PMCID: PMC9031365
- DOI: 10.3390/v14040781
Longitudinal Survey of Coronavirus Circulation and Diversity in Insectivorous Bat Colonies in Zimbabwe
Abstract
Background: Studies have linked bats to outbreaks of viral diseases in human populations such as SARS-CoV-1 and MERS-CoV and the ongoing SARS-CoV-2 pandemic.
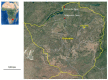
Methods: We carried out a longitudinal survey from August 2020 to July 2021 at two sites in Zimbabwe with bat-human interactions: Magweto cave and Chirundu farm. A total of 1732 and 1866 individual bat fecal samples were collected, respectively. Coronaviruses and bat species were amplified using PCR systems.
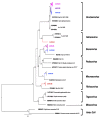
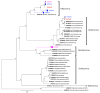
Results: Analysis of the coronavirus sequences revealed a high genetic diversity, and we identified different sub-viral groups in the Alphacoronavirus and Betacoronavirus genus. The established sub-viral groups fell within the described Alphacoronavirus sub-genera: Decacovirus, Duvinacovirus, Rhinacovirus, Setracovirus and Minunacovirus and for Betacoronavirus sub-genera: Sarbecoviruses, Merbecovirus and Hibecovirus. Our results showed an overall proportion for CoV positive PCR tests of 23.7% at Chirundu site and 16.5% and 38.9% at Magweto site for insectivorous bats and Macronycteris gigas, respectively.
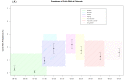
Conclusions: The higher risk of bat coronavirus exposure for humans was found in December to March in relation to higher viral shedding peaks of coronaviruses in the parturition, lactation and weaning months of the bat populations at both sites. We also highlight the need to further document viral infectious risk in human/domestic animal populations surrounding bat habitats in Zimbabwe.
Keywords: Zimbabwe; bat coronavirus (Bt CoVs); genetic diversity; human–bat interaction; reproductive phenology.
Conflict of interest statement
We declare that we have no conflict of interest.
Figures

References
-
- ICTV Positive Sense RNA Viruses: Coronaviridae. 2020. [(accessed on 1 December 2021)]. VIRUS Taxon. Available online: https://talk.ictvonline.org/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

