Dual Role of HIV-1 Envelope Signal Peptide in Immune Evasion
- PMID: 35458538
- PMCID: PMC9030904
- DOI: 10.3390/v14040808
Dual Role of HIV-1 Envelope Signal Peptide in Immune Evasion
Abstract
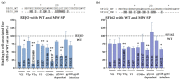
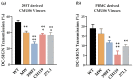
HIV-1 Env signal peptide (SP) is an important contributor to Env functions. Env is generated from Vpu/Env encoded bicistronic mRNA such that the 5' end of Env-N-terminus, that encodes for Env-SP overlaps with 3' end of Vpu. Env SP displays high sequence diversity, which translates into high variability in Vpu sequence. This study aimed to understand the effect of sequence polymorphism in the Vpu-Env overlapping region (VEOR) on the functions of two vital viral proteins: Vpu and Env. We used infectious molecular clone pNL4.3-CMU06 and swapped its SP (or VEOR) with that from other HIV-1 isolates. Swapping VEOR did not affect virus production in the absence of tetherin however, presence of tetherin significantly altered the release of virus progeny. VEOR also altered Vpu's ability to downregulate CD4 and tetherin. We next tested the effect of these swaps on Env functions. Analyzing the binding of monoclonal antibodies to membrane embedded Env revealed changes in the antigenic landscape of swapped Envs. These swaps affected the oligosaccharide composition of Env-N-glycans as shown by changes in DC-SIGN-mediated virus transmission. Our study suggests that genetic diversity in VEOR plays an important role in the differential pathogenesis and also assist in immune evasion by altering Env epitope exposure.
Keywords: DC-SIGN; HIV-1; Vpu; antibodies; envelope; glycosylation; tetherin antagonism; transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Vermeire J., Roesch F., Sauter D., Rua R., Hotter D., Van Nuffel A., Vanderstraeten H., Naessens E., Iannucci V., Landi A., et al. HIV Triggers a cGAS-Dependent, Vpu- and Vpr-Regulated Type I Interferon Response in CD4+ T Cells. Cell Rep. 2016;17:413–424. doi: 10.1016/j.celrep.2016.09.023. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

