Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern: A Perspective for Emerging More Transmissible and Vaccine-Resistant Strains
- PMID: 35458557
- PMCID: PMC9029021
- DOI: 10.3390/v14040827
Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern: A Perspective for Emerging More Transmissible and Vaccine-Resistant Strains
Abstract
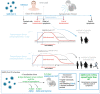
Novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOC) are constantly threatening global public health. With no end date, the pandemic persists with the emergence of novel variants that threaten the effectiveness of diagnostic tests and vaccines. Mutations in the Spike surface protein of the virus are regularly observed in the new variants, potentializing the emergence of novel viruses with different tropism from the current ones, which may change the severity and symptoms of the disease. Growing evidence has shown that mutations are being selected in favor of variants that are more capable of evading the action of neutralizing antibodies. In this context, the most important factor guiding the evolution of SARS-CoV-2 is its interaction with the host's immune system. Thus, as current vaccines cannot block the transmission of the virus, measures complementary to vaccination, such as the use of masks, hand hygiene, and keeping environments ventilated remain essential to delay the emergence of new variants. Importantly, in addition to the involvement of the immune system in the evolution of the virus, we highlight several chemical parameters that influence the molecular interactions between viruses and host cells during invasion and are also critical tools making novel variants more transmissible. In this review, we dissect the impacts of the Spike mutations on biological parameters such as (1) the increase in Spike binding affinity to hACE2; (2) bound time for the receptor to be cleaved by the proteases; (3) how mutations associate with the increase in RBD up-conformation state in the Spike ectodomain; (4) expansion of uncleaved Spike protein in the virion particles; (5) increment in Spike concentration per virion particles; and (6) evasion of the immune system. These factors play key roles in the fast spreading of SARS-CoV-2 variants of concern, including the Omicron.
Keywords: SARS-CoV-2 variants; sensitivity to antisera; transmissibility; viral load.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity.Semin Immunol. 2021 Jun;55:101533. doi: 10.1016/j.smim.2021.101533. Epub 2021 Nov 20. Semin Immunol. 2021. PMID: 34836774 Free PMC article. Review.
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
Research progress in spike mutations of SARS-CoV-2 variants and vaccine development.Med Res Rev. 2023 Jul;43(4):932-971. doi: 10.1002/med.21941. Epub 2023 Mar 16. Med Res Rev. 2023. PMID: 36929527 Review.
-
Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil.Microbiol Spectr. 2022 Feb 23;10(1):e0236621. doi: 10.1128/spectrum.02366-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196783 Free PMC article.
Cited by
-
Omicron BA.2 Lineage, the "Stealth" Variant: Is It Truly a Silent Epidemic? A Literature Review.Int J Mol Sci. 2022 Jun 30;23(13):7315. doi: 10.3390/ijms23137315. Int J Mol Sci. 2022. PMID: 35806320 Free PMC article. Review.
-
Low-Entropy Hydration Shells at the Spike RBD's Binding Site May Reveal the Contagiousness of SARS-CoV-2 Variants.Biomolecules. 2023 Nov 7;13(11):1628. doi: 10.3390/biom13111628. Biomolecules. 2023. PMID: 38002310 Free PMC article.
-
COVID-19-related deaths: a 2-year inter-wave comparison of mortality data from Germany.Infection. 2023 Aug;51(4):1147-1152. doi: 10.1007/s15010-023-01982-4. Epub 2023 Jan 24. Infection. 2023. PMID: 36690889 Free PMC article.
-
Long covid outcomes at one year after mild SARS-CoV-2 infection: nationwide cohort study.BMJ. 2023 Jan 11;380:e072529. doi: 10.1136/bmj-2022-072529. BMJ. 2023. PMID: 36631153 Free PMC article.
-
Mutational dynamics of SARS-CoV-2: Impact on future COVID-19 vaccine strategies.Heliyon. 2024 Apr 25;10(9):e30208. doi: 10.1016/j.heliyon.2024.e30208. eCollection 2024 May 15. Heliyon. 2024. PMID: 38707429 Free PMC article. Review.
References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 3 February 2022)]. Available online: https://covid19.who.int/
-
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

